Hi Seb,
First of all, wow!. Pretty neat job of benchmarking here! I think you’ve taken this farther I’ve ever tried :D.
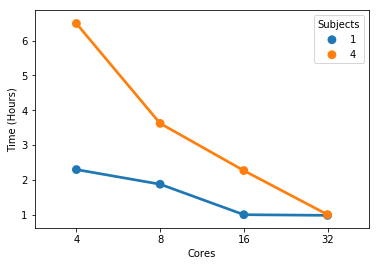
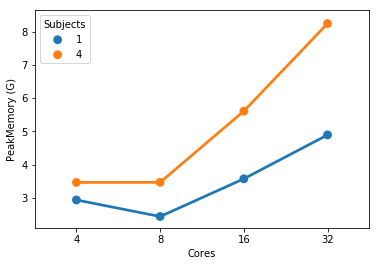
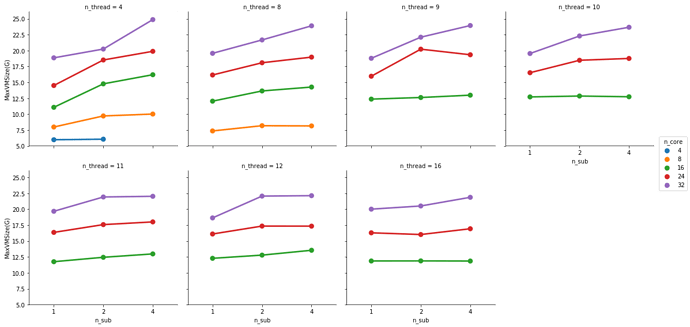
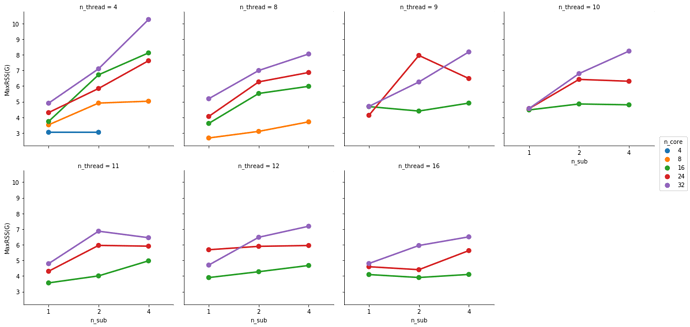
At 32 cores, 1 sample and 4 samples take the same amount of completion time but the 4 sample instance uses more memory (not sure these memory numbers make too much sense but that’s what the scheduler tells me).
So there seems to be some minimal runtime that I can’t get below by throwing more cores at it - possibly because a portion of the pipeline has sequential jobs.
Yes, there is a limit and the reason is what you mentioned: sequential jobs.
So in a 32 cores and 64G memory instance I have some room left to run additional samples in parallel.
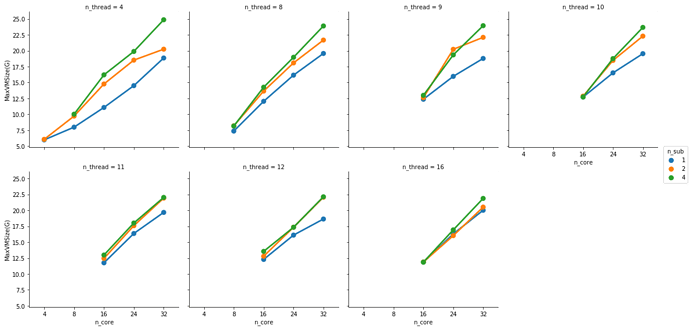
This is partially true. FMRIPREP is still a bit inefficient when allocating memory. Even though we made a great job at keeping physical memory (typically referred to as RSS) low (we just need to look at your graph), FMRIPREP has some trouble with the virtual memory fingerprint.
If your system is very flexible about overcommitting memory this will probably not surface as a problem for you. But typically, HPCs will set hard limits and the OOM killer will kick in.
So, to really find the hard limit on your settings you will need to add more samples to your tests and look for the output logs of the scheduler. When FMRIPREP attempts to get pass your 64GB limit you will probably have some warnings. At some number of samples, the kernel will kill your job. However, given that you won’t go over 32 cores and seeing that the runtime is the same for 1 or 4 subjects at 32, maybe 32 is the maximum number of parallelizable tasks regardless the number of samples. If that’s correct, then the memory issues should not appear for you.
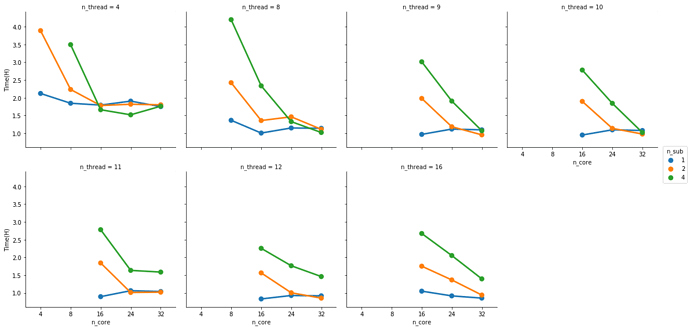
Possibly the number of threads each task can use would also have an influence here. But before I run more tests I’d like to ask if you have found some rule of thumb for running efficient instances. Something like: 3 samples at 16 cores and 32G of memory take an hour and have a consistently high CPU and RAM usage.
I generally run 1 subject per node. Since we started to face memory issues I haven’t tried more, but I’m likely under-using the HPC resource here. On our case, the memory allocation blows up after 10 parallel jobs (--n_procs 10).
By augmenting the number of threads per task (--omp_nthreads) you may speed up the process a lot (and also hit memory issues). But there are a couple of bottlenecks that will scale very well with the number of threads per task. So you are in the right path and your intuitions are impecable.
I’m very looking forward to hearing where you got with this 
Cheers,
Oscar

 ), and I need to preprocess 110 subjects, but can’t afford to wait ~100 days to start second levels.
), and I need to preprocess 110 subjects, but can’t afford to wait ~100 days to start second levels.