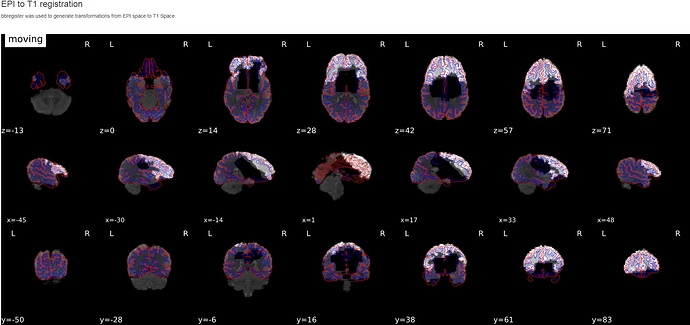
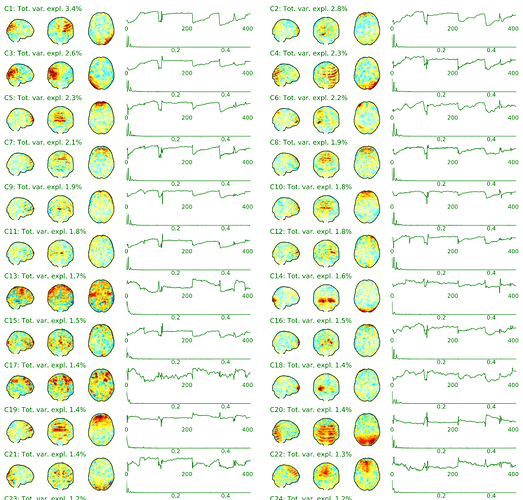
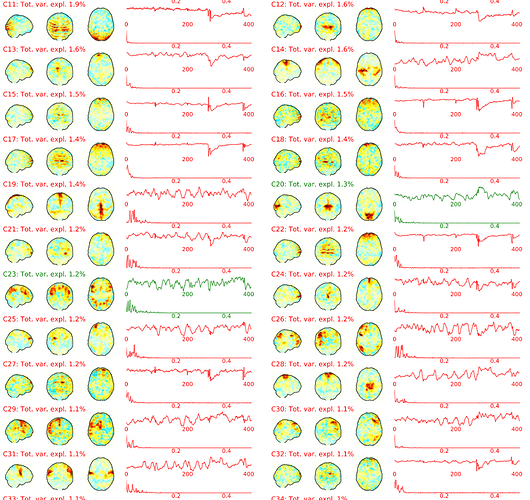
Hi there,
I’ve started using fmriprep very recently to preprocess my data on our lab’s Linux cluster. I am encountering several issues and I am trying to determine if they are due to bugs in the pipeline or to computing issues related to the clusters.
So far, I’ve mainly tried to only run one subject at a time (or max 1 subject per node).
I am using FMRIPREP version: 1.0.6-2, from a singularity container, and my fmriprep command is as follows:
singularity run /home/ccharpen/fmriprep/poldracklab_fmriprep_latest-2018-01-30-2863112d2d8d.img -w /home/ccharpen/ObsLearn/prepdata/work /home/ccharpen/ObsLearn/rawdata/rawBIDS /home/ccharpen/ObsLearn/prepdata/fmriprep participant --participant-label 007 --ignore slicetiming --use-aroma --ignore-aroma-denoising-errors --fs-license-file /home/ccharpen/fmriprep/license.txt
So far, out of 7 subjects, one has succeeded on the first try. For all the others, it’s either crashing during freesurfer surface reconstruction or during ICA-AROMA for one or more functional runs (see error messages below).
At first my guess was that it was maybe a memory error since those processes seem to be the heaviest in terms of memory usage. Also our HPcluster is set up to kill jobs when they go over the requested memory for that job (which I had specified at 6GB); but when trying to re-run them with either 24GB or 32GB, some subjects ran but some others still crashed - which makes me think there is more to it than memory…
Any thoughts?? Let me know if you need more information!
Thanks,
Caroline
Freesurfer recon-all error:
Applying LTAtransformInterp (resample_type 1)
writing to /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/mri/T2.prenorm.mgz…mri_normalize -sigma 0.5 -nonmax_suppress 0 -min_dist 1 -aseg /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/mri/aseg.presurf.mgz -surface /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/surf/rh.white identity.nofile -surface /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/surf/lh.white identity.nofile /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/mri/T2.prenorm.mgz /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/mri/T2.norm.mgz
mghRead(/home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/mri/T2.prenorm.mgz): could not read 262144 bytes at slice 150
using Gaussian smoothing of bias field, sigma=0.500
disabling nonmaximum suppression
retaining points that are at least 1.000mm from the boundary
using segmentation for initial intensity normalization
reading from /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/mri/T2.prenorm.mgz…
No such file or directory
mri_normalize: could not open source file /home/ccharpen/ObsLearn/prepdata/fmriprep/freesurfer/sub-004/mri/T2.prenorm.mgz
No such file or directory
Linux master 3.13.0-109-generic #156-Ubuntu SMP Wed Feb 8 16:09:17 UTC 2017 x86_64 x86_64 x86_64 GNU/Linuxrecon-all -s sub-004 exited with ERRORS at Thu Mar 22 10:21:51 UTC 2018
ICA-AROMA error:
Traceback (most recent call last):
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/nipype/pipeline/plugins/multiproc.py”, line 68, in run_node
result[‘result’] = node.run(updatehash=updatehash)
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/nipype/pipeline/engine/nodes.py”, line 487, in run
result = self._run_interface(execute=True)
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/nipype/pipeline/engine/nodes.py”, line 571, in _run_interface
return self._run_command(execute)
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/nipype/pipeline/engine/nodes.py”, line 650, in _run_command
result = self._interface.run(cwd=outdir)
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/nipype/interfaces/base/core.py”, line 516, in run
runtime = self._run_interface(runtime)
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/interfaces/report_base.py”, line 54, in _run_interface
ReportCapableInterface, self)._run_interface(runtime)
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/nipype/interfaces/base/core.py”, line 1023, in _run_interface
self.raise_exception(runtime)
File “/usr/local/miniconda/lib/python3.6/site-packages/niworkflows/nipype/interfaces/base/core.py”, line 960, in raise_exception
).format(**runtime.dictcopy()))
RuntimeError: Command:
ICA_AROMA.py -den nonaggr -i /home/ccharpen/ObsLearn/prepdata/work/fmriprep_wf/single_subject_007_wf/func_preproc_task_ObsLearn_run_04_wf/bold_confounds_wf/ica_aroma_wf/smooth/vol0000_xform-00000_merged_smooth.nii.gz -meldir /home/ccharpen/ObsLearn/prepdata/work/fmriprep_wf/single_subject_007_wf/func_preproc_task_ObsLearn_run_04_wf/bold_confounds_wf/ica_aroma_wf/melodic -mc /home/ccharpen/ObsLearn/prepdata/work/fmriprep_wf/single_subject_007_wf/func_preproc_task_ObsLearn_run_04_wf/bold_hmc_wf/normalize_motion/motion_params.txt -o /home/ccharpen/ObsLearn/prepdata/work/fmriprep_wf/single_subject_007_wf/func_preproc_task_ObsLearn_run_04_wf/bold_confounds_wf/ica_aroma_wf/ica_aroma
Standard output:------------------------------- RUNNING ICA-AROMA -------------------------------
--------------- ‘ICA-based Automatic Removal Of Motion Artifacts’ ---------------Warning! Please check whether the determined TR (of 1.0s) is correct!
Step 1) MELODIC
- The existing/specified MELODIC directory will be used.
- The MELODIC directory does not contain the required ‘stats’ folder. Mixture modeling on the Z-statistical maps will be run.
Step 2) Automatic classification of the components- registering the spatial maps to MNI
- extracting the CSF & Edge fraction features
- The spatial map of component 1 is empty. Please check!
- The spatial map of component 2 is empty. Please check!
- The spatial map of component 3 is empty. Please check!
- The spatial map of component 4 is empty. Please check!
- The spatial map of component 5 is empty. Please check!
- The spatial map of component 6 is empty. Please check!
- The spatial map of component 7 is empty. Please check!
- The spatial map of component 8 is empty. Please check!
- The spatial map of component 9 is empty. Please check!
- The spatial map of component 10 is empty. Please check!
- The spatial map of component 11 is empty. Please check!
- The spatial map of component 12 is empty. Please check!
- The spatial map of component 13 is empty. Please check!
- The spatial map of component 14 is empty. Please check!
- The spatial map of component 15 is empty. Please check!
- The spatial map of component 16 is empty. Please check!
- The spatial map of component 17 is empty. Please check!
- The spatial map of component 18 is empty. Please check!
- The spatial map of component 19 is empty. Please check!
- The spatial map of component 20 is empty. Please check!
- The spatial map of component 21 is empty. Please check!
- The spatial map of component 22 is empty. Please check!
- The spatial map of component 23 is empty. Please check!
- The spatial map of component 24 is empty. Please check!
- The spatial map of component 25 is empty. Please check!
- The spatial map of component 26 is empty. Please check!
- The spatial map of component 27 is empty. Please check!
- The spatial map of component 28 is empty. Please check!
- The spatial map of component 29 is empty. Please check!
- The spatial map of component 30 is empty. Please check!
- The spatial map of component 31 is empty. Please check!
- The spatial map of component 32 is empty. Please check!
- The spatial map of component 33 is empty. Please check!
- The spatial map of component 34 is empty. Please check!
- The spatial map of component 35 is empty. Please check!
- The spatial map of component 36 is empty. Please check!
- The spatial map of component 37 is empty. Please check!
- The spatial map of component 38 is empty. Please check!
- The spatial map of component 39 is empty. Please check!
- The spatial map of component 40 is empty. Please check!
- The spatial map of component 41 is empty. Please check!
- The spatial map of component 42 is empty. Please check!
- The spatial map of component 43 is empty. Please check!
- The spatial map of component 44 is empty. Please check!
- The spatial map of component 45 is empty. Please check!
- The spatial map of component 46 is empty. Please check!
- The spatial map of component 47 is empty. Please check!
- The spatial map of component 48 is empty. Please check!
- The spatial map of component 49 is empty. Please check!
- The spatial map of component 50 is empty. Please check!
- The spatial map of component 51 is empty. Please check!
- The spatial map of component 52 is empty. Please check!
- The spatial map of component 53 is empty. Please check!
- The spatial map of component 54 is empty. Please check!
- The spatial map of component 55 is empty. Please check!
- The spatial map of component 56 is empty. Please check!
- The spatial map of component 57 is empty. Please check!
- The spatial map of component 58 is empty. Please check!
- The spatial map of component 59 is empty. Please check!
- The spatial map of component 60 is empty. Please check!
- The spatial map of component 61 is empty. Please check!
- The spatial map of component 62 is empty. Please check!
- The spatial map of component 63 is empty. Please check!
- The spatial map of component 64 is empty. Please check!
- The spatial map of component 65 is empty. Please check!
- The spatial map of component 66 is empty. Please check!
- The spatial map of component 67 is empty. Please check!
- The spatial map of component 68 is empty. Please check!
- The spatial map of component 69 is empty. Please check!
- The spatial map of component 70 is empty. Please check!
- The spatial map of component 71 is empty. Please check!
- The spatial map of component 72 is empty. Please check!
- The spatial map of component 73 is empty. Please check!
- The spatial map of component 74 is empty. Please check!
- The spatial map of component 75 is empty. Please check!
- The spatial map of component 76 is empty. Please check!
- The spatial map of component 77 is empty. Please check!
- The spatial map of component 78 is empty. Please check!
- The spatial map of component 79 is empty. Please check!
- The spatial map of component 80 is empty. Please check!
- The spatial map of component 81 is empty. Please check!
- The spatial map of component 82 is empty. Please check!
- The spatial map of component 83 is empty. Please check!
- The spatial map of component 84 is empty. Please check!
- The spatial map of component 85 is empty. Please check!
- The spatial map of component 86 is empty. Please check!
- The spatial map of component 87 is empty. Please check!
- The spatial map of component 88 is empty. Please check!
- The spatial map of component 89 is empty. Please check!
- The spatial map of component 90 is empty. Please check!
- The spatial map of component 91 is empty. Please check!
- The spatial map of component 92 is empty. Please check!
- The spatial map of component 93 is empty. Please check!
- The spatial map of component 94 is empty. Please check!
- The spatial map of component 95 is empty. Please check!
- The spatial map of component 96 is empty. Please check!
- The spatial map of component 97 is empty. Please check!
- The spatial map of component 98 is empty. Please check!
- The spatial map of component 99 is empty. Please check!
- The spatial map of component 100 is empty. Please check!
- The spatial map of component 101 is empty. Please check!
- The spatial map of component 102 is empty. Please check!
- The spatial map of component 103 is empty. Please check!
- The spatial map of component 104 is empty. Please check!
- extracting the Maximum RP correlation feature
- extracting the High-frequency content feature
- classification
Standard error:
Traceback (most recent call last):
File “/opt/ICA-AROMA/ICA_AROMA.py”, line 211, in
motionICs = aromafunc.classification(outDir, maxRPcorr, edgeFract, HFC, csfFract)
File “/opt/ICA-AROMA/ICA_AROMA_functions.py”, line 487, in classification
if len(motionICs) != 0:
TypeError: len() of unsized object