Hi All,
When running fMRI prep I’m getting the following error:
“mismatched qform/sform matrixes”
When running the Bids validator I’m getting the following error:
sform_code and qform_code in the image header are 0.
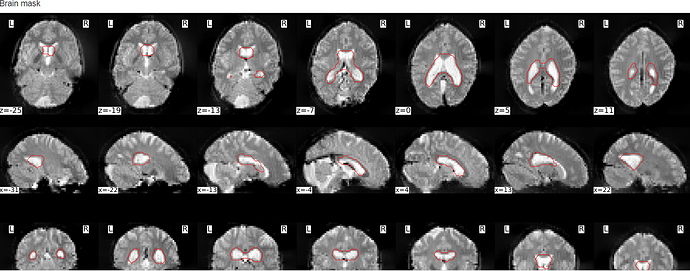
When looking at the log file this is the output I’m getting for the brain mask:
Is there something I can do to get fmriprep running even though these codes are missing?
Thank you very much for your help,
Sebastian
The headers are corrupted and thus the orientation information cannot be trusted. I recommend converting the data from dicoms or par/rec using latest version of dcm2niix https://github.com/rordenlab/dcm2niix.
@ChrisGorgolewski I have now used dcm2niix as implemented in nipype (http://nipype.readthedocs.io/en/latest/interfaces/generated/nipype.interfaces.dcm2nii.html) and still receive the same error. Is there anything else that may cause this issue?
Not sure what’s going on. My best advice would be to open an issue at https://github.com/rordenlab/dcm2niix/issues
and provide example dicoms.
Okay thanks I will try that!
Hello,
I think the issue is with your pipeline, not dcm2niix. Please see the github Issue 197 for this image. The q-form’s quaternion in the NIfTI header needs to be converted to a matrix in order to compare to the s-form (which is stored natively as a matrix). In particular, I think you can see values that are +0.000000 for the s-form and -0.00000 for the q-form (and vice versa). This can cause precision errors. This is explaned here here for Python. For C, you would compare elements of these matrices with a tolerance for error. If you look at dcm2niix, you will see the C solution for this problem:
int isSameDouble (double a, double b) {
return (fabs (a - b) <= DBL_EPSILON);
}
For Python, I think you want to use numpy.isclose. With regards to the tolerance, bear in mind that both the s-form and q-form are stored as 32-bit floats in NIfTI1.
But would this explain why BIDS Validator detects qform_code and sform_code to be set to zero?
Well dcm2niix fills both the sform and qform. So either the pipeline corrupted it or the message needs to be changed.
@Sebastian - could you convert your dicom files using the latest version of dcm2niix downloaded from github, check if the output niftis have qform_code and sform_form set to non zero (using for example fslhd), and finally see if the BIDS validator still complains about sform/qform_code? Thx in advance.
@ChrisGorgolewski - I have now converted the data using the latest version on the command line and the qform and sform_code are both set to 1. I get the same sform_code and qform_code when running the conversion via nipype. The BIDS validator now does not give me any errors anymore.
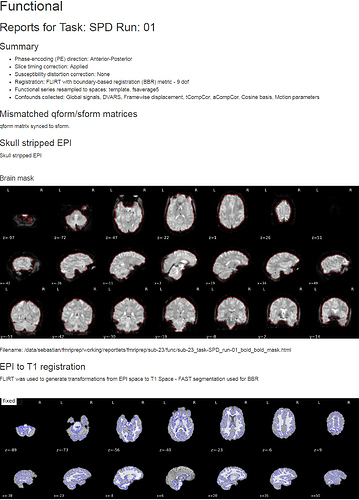
However, when running fMRIprep the message still pops up, but the preprocessing for the functional scan now seems to work fine:
So can I just ignore the error message?
The message is just a notification. We do not consider it an error, but it was felt that people might want to know, as its unexpected appearance may indicate an issue with their data.
If you want to see what fmriprep sees, you can use nibabel:
import nibabel as nb
img = nb.load(fname)
print(img.get_qform(coded=True))
print(img.get_sform(coded=True))
If you’re still getting that error after syncing the matrices, it may actually be just a reporting glitch, and removing your output directory ought to clear it up.