Hello TDT users,
I have 16 beta images for each of the 3 conditions across four runs per subject, which were used as the input for individual’s searchlight analysis.
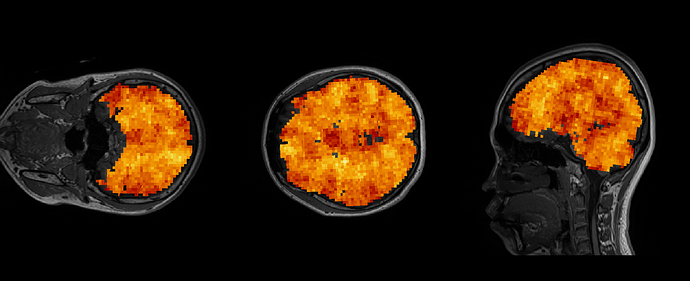
I predicted that some decodable regions will show positive value in the file ‘res_accuracy_minus_chance.nii’, but all voxels in the whole brain show above chance level in TDT results unexpectedly. (I uploaded the file as example.)
So, I wonder if this result is valid, or if I set wrong parameters for decoding analysis.
The parameters that I used is as below:
cfg = decoding_defaults(cfg);
cfg.testmode = 0;
cfg.analysis = decoding_type;
cfg.decoding.method = ‘classification_kernel’;
cfg.decoding.software = ‘libsvm’;
cfg.decoding.train.classification.model_parameters = ‘-s 0 -t 0 -c 1 -b 0 -q’;
cfg.software = spm(‘ver’);
cfg.results.dir = output_dir;
cfg.searchlight.radius = 12;
cfg.searchlight.unit = ‘mm’;
cfg.files.mask = fullfile(mask_dir);
cfg.files.name = alldesign(:,1);
cfg.files.label = cat(1, alldesign{:,2});
cfg.files.chunk = cat(1, alldesign{:,3});
%(I checked that the beta images are allocated well in the design matrix.)
cfg.design = make_design_cv(cfg);
cfg.results.output = {‘accuracy’,‘accuracy_minus_chance’};
results = decoding(cfg);
Any feedback would be great, Thanks!
~ Taehyun Yoo