That is real weird I’ll have to call Chris on this because he knows linux better than me (I’m a Mac person sorry…). He should reply to you later today. meanwhile can you just check that there is no space sign in any of your directories. cheers mich
Hi Max,
It’s weird, I don’t remember having this error. The only things I can see from here are either a permission problem or an empty input directory (either your input directory or the BCBToolkit/Tools/extraFiles/tracks)
Have you tried with your own data?
Cheers,
Chris.
Hi Chris,
neither the input direcotry nor the tracks directory were empty. I also tried with my own data. I eventually got it to work using a coworkers Macbook …
Cheers, Max
Hi Chris and Michel
I’m working wiht BCBToolkit and have a few questions.
I try to perform a Kruskall-Wallis analysis with Bonferonni-Holm correction. I don’t understand the output files in my studies.
In fact, my Kruskal_pvalues.csv file shows that my 54 clusters have a pvalue < 0.05 and that the kw_holm is also < 0.05 that means all my clusters survived the BH correction.
However the last 3 columns (pval, stat and holm) show N/A and I don’t understand why. Does that mean the posthoc test failed ?
In previous works (in which I believed I have understood something  ) I thought that values of the voxel should correspond to the value in the csv file so we can filter voxel < 0.05 to identify significant voxel. (and one consequence was that if no voxel survived the BH correction, all the voxels of the file were at an intensity of 1 so there was a big white square). But my nifti files are curious (I open them in MRIcron or MRIcroGL) : indeed the kruskall_holm_clusters.nii.gz should indicate BH corrected pvalue after the KW test, and so all my voxel values should be < 0.05 but the map show voxel with 0.99 intensity level. It is the same for the kruskal_cluster.nii.gz where the value of the voxels do not correspond to the value in the csv file .But I had also curious results with voxels value at 0 but with clearly a white voxel on the screen corresponding to significant clusters and the voxel should have an intensity of 1…). Concerning the cluster.nii and cluster_holm.nii I don’t even know where to find the value…
) I thought that values of the voxel should correspond to the value in the csv file so we can filter voxel < 0.05 to identify significant voxel. (and one consequence was that if no voxel survived the BH correction, all the voxels of the file were at an intensity of 1 so there was a big white square). But my nifti files are curious (I open them in MRIcron or MRIcroGL) : indeed the kruskall_holm_clusters.nii.gz should indicate BH corrected pvalue after the KW test, and so all my voxel values should be < 0.05 but the map show voxel with 0.99 intensity level. It is the same for the kruskal_cluster.nii.gz where the value of the voxels do not correspond to the value in the csv file .But I had also curious results with voxels value at 0 but with clearly a white voxel on the screen corresponding to significant clusters and the voxel should have an intensity of 1…). Concerning the cluster.nii and cluster_holm.nii I don’t even know where to find the value…
Is it possible that the value of the voxel, if too low or with to many decimals could be misinterpreted by a software of visualization ?
Moreover I’m not sure to understand the difference between the nifti files very well when using the KW test with BH correction:
- Does the clusters.nii file show the posthoc results for the clusters which passed the KW test ?
- the kruskal_cluster.nii indicates pvalues ot the KW test before BH
- the kruskal_holm_clusters indicates pvalue of the BH correction after KW
- but what is the cluster_holm.nii when using KW and BH correction ? Does it show the significant cluster that passed the BH correction after KW test and the BH after post hoc test so the very end of the process ?
Sorry for this long question and thank you in advance.
Best regards
Fabien
Hi Fabien,
Sorry you’re having troubles with the toolkit :s.
So, the kruskal_pvalues.csv should contain the results of the tests. And if they have N/A yes, the tests went wrong. There should be another file called ‘test_warn.csv’ in case some warnings/errors were raised by the function performing the stats. You might have more info about what went wrong in there. You can send the messages or the file to me to have a look if it does not help.
The rest of the weird results is likely due to this error. The script tries to fill up the images but does not have the values so the images are broken.
In the nifti images, you should have 1 - pvalue or 1 - corrected_pvalue (apparently that’s a standard way to add the stats in an image). That’s why you can have .99, which is a significant cluster in that case.
Oh, and if there is Kruskal or kw in the name it refers to the kruskal wallis test only. Then cluster and cluster_holm are the posthoc stats between the two groups you selected (if they pass the kruskal wallis).
Hope it helps.
Best regards,
Chris.
Dear Chris and Michel,
I’m trying to run the Normalization tool but I’m getting the following error:
“Cannot open volume /home/edu_asus/Documentos/Grant/NIH/bcbtoolkit/Lesion/subj01AXT1.nii* for reading!”
I don’t know why it is looking for the file “subj01AXT1.nii” in the Lesion folder.
Running just with the T1 image it is ok. The problem is when I include the Lesion folder.
The lesion is in the the native space of the T1 image. I have also tried to change both images to the MNI space using:
flirt -interp nearestneighbour -in subj01lesion.nii.gz -ref $FSLDIR/data/standard/MNI152_T1_1mm_brain.nii.gz -out wsubj01lesion_lesion.nii -usesqform -applyxfm
flirt -interp nearestneighbour -in subj01AXT1.nii.gz -ref $FSLDIR/data/standard/MNI152_T1_1mm_brain.nii.gz -out wsubj01lesion_lesion.nii -usesqform -applyxfm
But I’ve got the same error. Please, how can I fix it?
I’m using Ubuntu 20.04. Follow attached my log file.
Sincerely,
Eduardo
logNormalisation.txt (3.0 KB)
Hi Eduardo,
This one is easy.
The lesion and the T1 image must have the exact same name for the normalisation to work.
We did that in order for users to be able to process several patients at the same time.
I should update our userguide!
Http://www.bcblab.com/BCB/Normalisation.html
Kind regards
Mich
Hi Dr Mich,
Sorry about that. Now I could see this information in the userguide:
" **exact same name as the T1 images used above "
Thank you!
My pleasure hope you will have fun with BCBtoolkit!
Hi Chris and Michel,
I’m attempting to use AnaCOM2 on the disconnectome images generated from the Disconnectome Map tool but got following message in terminal:
“Exception in thread “AWT-EventQueue-0” java.lang.ClassCastException: sun.awt.image.BufImgSurfaceData cannot be cast to sun.java2d.xr.XRSurfaceData”
It seems like the analysis is still running so I am unsure whether I should be concerned? For context, I am processing the data on a linux HPC desktop. I have 26 participants with disconnectome images and it has been running for about 2 days now.
Regards,
Abbie
Hi Abbie,
This is an exception thrown by the java GUI so, it will not affect the results of the analysis. I have seen some exceptions coming up when my screen turns off or things like that but the program is still running so it should be fine :).
Regards,
Chris
Dear Chris and Michel,
I am attempting to use the Disconnectome Maps feature of the BCBToolKit, using the additional Package X dataset (which I believe I have correctly added to the ToolKit’s folder). I am trying to calculate disconnectome maps using the ToolKit, and while this seems to have worked, it seems that some of the disconnectome map files I get as a result are empty (i.e., the BCBToolKit creates them, but there are just 0s in the nifti file).
To try to fix this, I have resliced some of my lesioned files to ensure that they were using the BCBToolKit MNI template (rather than the SPM one that I originally had, as per your advice in an earlier thread) in case this was the problem, but it does not seem to have helped. After having run some of these resliced nifti files through the ToolKit, I get nice looking disconnectome maps for some of my participants, but not all – for some of my participants, the resulting disconnectome map nifti file is still empty. Unfortunately, I’m not able to attach log files to illustrate as I’m a new user, but can do so by email if that helps!
Do you have any further ideas on what might be causing this? Also, while I know the ToolKit’s documentation states the lesioned nifti files should be binarised, not all of my lesioned nifti files are and some of these have still produced disconnectome maps. Is this possibly problematic?
Apologies for the lengthy description – I hope it’s not too confusing! Let me know if I can clarify my problem further. And thank you very much for developing the ToolKit and for having this forum for questions!!
Thanks very much,
Nadene
Hi Nadene,
So, yes the images have to be in the same space as the tractographies used.
For the images not showing anything in the output, like that I can see a couple of explanations. If your lesions are outside of the tractographies there won’t be any fibre crossing the lesions. It can happen with erroneous registration, or maybe some lesions that are at the very surface of the cortex or small lesions outside of what’s tracked with the tractography.
If you can send me a couple of lesion masks that did not work and I can have a look if you want.
Concerning the non-binary masks, it could potentially be a problem if the values are between 0 and 1 or even negative, I haven’t tested it but, I am guessing track_vis is taking all the voxels above 1 internally. That’s why we usually advise users to binarize the masks.
Regards,
Chris.
Hi Chris,
I am trying to do disconnectome mapping on a Linux Ubuntu platform and have the same problem as Max.
Data is supposedly properly written (but it only takes a sec, so probably nothing was computed… )
The log file is empty.
Has there been a fix for this for linux machines?
Best regards,
Julian
Hi Julian,
Sorry you’re having issues with the BCBtoolkit.
What version of Ubuntu are you using?
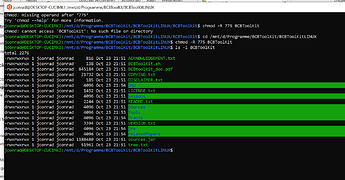
Have you checked the permissions of the BCBtoolkit folder? You can do it by using ”cd” into the parent folder of the BCBtoolkit and then use ”ls -l BCBToolkit”
You can try
”chmod -R 775 BCBToolkit” to change the permissions of the bcbtoolkit folder.
Hi Chris,
thanks for the quick reply. To make matters more complicated, I created a Linux workspace with WSL2 on my Windows 10 machine. Ubuntu is version 2004.20221.825.0.
these are the permissions,but so far it is not working.
Any ideas?
-rwxrwxrwx 1 jconrad jconrad 816 Oct 23 21:51 ACKNOWLEDGEMENT.txt
-rwxrwxrwx 1 jconrad jconrad 138 Oct 23 21:51 BCBToolKit.sh
-rwxrwxrwx 1 jconrad jconrad 845184 Oct 23 21:51 BCBtoolkit_doc.pdf
-rwxrwxrwx 1 jconrad jconrad 21732 Oct 23 21:51 COPYING.txt
-rwxrwxrwx 1 jconrad jconrad 185 Oct 23 21:51 DISCLAIMER.txt
drwxrwxrwx 1 jconrad jconrad 4096 Oct 23 21:54 JHU
-rwxrwxrwx 1 jconrad jconrad 1452 Oct 23 21:51 LICENSE.txt
drwxrwxrwx 1 jconrad jconrad 4096 Oct 23 21:51 Lesions
-rwxrwxrwx 1 jconrad jconrad 2244 Oct 23 21:51 README.txt
drwxrwxrwx 1 jconrad jconrad 4096 Oct 23 21:51 Sources
drwxrwxrwx 1 jconrad jconrad 4096 Oct 23 21:53 Tools
drwxrwxrwx 1 jconrad jconrad 4096 Oct 23 21:54 Tracts
-rwxrwxrwx 1 jconrad jconrad 3394 Oct 23 21:51 VERSION.txt
drwxrwxrwx 1 jconrad jconrad 4096 Oct 23 21:54 jre
drwxrwxrwx 1 jconrad jconrad 4096 Oct 23 21:54 relatedPapers
-rwxrwxrwx 1 jconrad jconrad 1388480 Oct 23 21:51 sources.jar
-rwxrwxrwx 1 jconrad jconrad 51961 Oct 23 21:51 tree.txt
Best regards,
Julian
Oh I never tried with wsl2. It’s still from a windows machine so I guess there could be different things going on in the background. Have you been able to install FSL and run its commands normally?
The BCBtoolkit is using fsl’s binaries contained in the package
Hey Chris,
so, after some tears I reestablished everything from scratch. I used Ubuntu 18.04 now and reinstalled fsl. With the newer version of ubuntu fsl was not running properly somehow. Now everything is running smoothly.
thanks for your help!
Best regards,
Julian
Hi BCBToolKit group,
I apologize for asking such a basic question but I’m having issues running the BCBToolKit.command script on my MBP (OS 11.6). My understanding is java is deprecated/is incompatible with newer mac operating systems. Is there a software version that is compatible with Big Sur or do I need to use a machine with an older OS if I want to use the BCB toolkit?
Thanks!
Frank
Hi Franck, I see you have withdrawn your questions, but you can find details here in any case.
Note for Catalina users
you might have come across difficulties for running BCBtoolkit and many other software.
We found an easy fix.
- Open your terminal and run the following command: sudo spctl --master-disable
You will need to provide your administrator password.
For the most recent version of McOSX you will have to authorize files one by one.