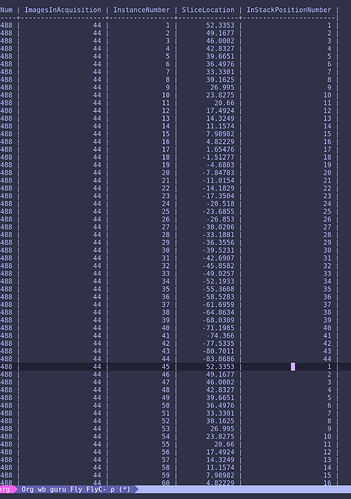
Hello,
I have a question regarding metadata extraction from DICOM files into *.json file associated with 4-D *.nii functional data file. According to BIDS specification (http://bids.neuroimaging.io/bids_spec.pdf page 11) dcm2nii extract BIDS compatible metadata. However, after conversion I discovered that onset values in the slice timing field in json file are not in seconds:
"SliceTiming": [ 0.00019523, 9.524e-05, 0.00019047, 9.047e-05, 0.00018571, 8.571e-05, 0.00018095, 8.095e-05, 0.00017619, 7.619e-05, 0.00017143, 7.143e-05, 0.00016666, 6.666e-05, 0.0001619, 6.19e-05, 0.00015714, 5.714e-05, 0.00015238, 5.238e-05, 0.00014762, 4.762e-05, 0.00014286, 4.285e-05, 0.00013809, 3.809e-05, 0.00013333, 3.333e-05, 0.00012857, 2.857e-05, 0.00012381, 2.381e-05, 0.00011905, 1.904e-05, 0.00011428, 1.428e-05, 0.00010952, 9.52e-06, 0.00010476, 4.76e-06, 0.0001, 0 ],
Other fields have correct units (e.g. "RepetitionTime": 2,). I can manually correct those values to fit in range [0, 2], however I am not sure if dcm2nii works properly. Previously I was told that slice order for this dataset was set to [ 1 : 2 : 41, 2 : 2 : 42 ] (in MATLAB / Octave notation). Data were acquired in 3T GE scanner. Those onset values suggest reverse order of acquisition.
I also tried to use dcm2niix, but apparently it doesn’t include slice timing in json extracted metadata.
Did anyone encounter similar issue? Should I reformat those values using script or use different tool to extract valid metadata?
Thanks,
Kamil Bonna