Is anyone working on an fmriprep equivalent for diffusion? Ideally in a BIDS-App ?
There’ve definitely been people interested in it, but I don’t know that things have materialized into a public repository anywhere.
If anybody does start working on this, we may want to coordinate to ensure that the anatomical preprocessing is consistent with fMRIPrep’s.
I’m writing it.
LoC this far: 0.
(now seriously, yes, Russ has asked a couple of times and I want to do it).
Here’s what i have so far: https://github.com/akeshavan/preAFQ/blob/master/preafq/run_1.py#L5 – right now the code is a little tangled to run with HBN data on AWS
and it generates a .json which can be rendered to http://anisha.pizza/preAFQ/#/report?url=https%3A%2F%2Fgist.githubusercontent.com%2Fakeshavan%2F89b8be9edf6a82d4a85745c7d61df0fe%2Fraw%2F1b5a342335ce6059a491fc1d2f25de44bae10dac%2Freport.json
Seems that @Ariel_Rokem (or @arokem) renabmed preAFQ to dmriprep (see https://github.com/nipy/dmriprep and announcement at https://mail.python.org/pipermail/neuroimaging/2018-November/001841.html)
Yep. We’ve uploaded it to a more prominent place and renamed it with the goal of moving this ahead and giving @oesteban a place to put his work 
Update: we’ve started the migration from akeshavan/preAFQ to nipy/dmriprep and the new link to the report is: http://nipy.org/dmriprep/#/report?url=https%3A%2F%2Fgist.githubusercontent.com%2Fakeshavan%2F89b8be9edf6a82d4a85745c7d61df0fe%2Fraw%2F1b5a342335ce6059a491fc1d2f25de44bae10dac%2Freport.json
if anyone has ideas of what other QC images/metrics we should compute, let us know!
Very beautiful work…!
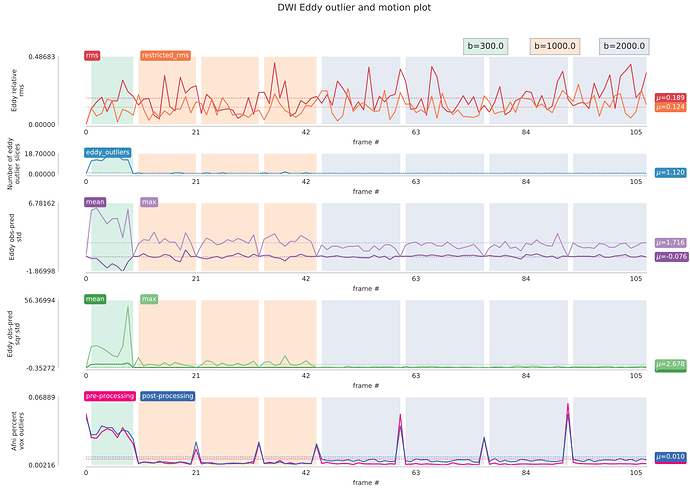
I’ve also been working on QC web page for our diffusion pipeline, but nothing fancy, just static images… But I basically check the same things you guys included in the dmriprep QC. In addition, I look at shell-specific tSNR images (we have multi-shell data) and eddy CNR maps. I also perform shell-specific outlier detection with 3dToutCount. These were inspired by Roelf et al 2016 paper (https://www.ncbi.nlm.nih.gov/pubmed/26520775). The outlier output is by frame, so I put them back together to produce combined Eddy rms/outliers and afni outliers like below;
I hope these will give you additional ideas for your report 
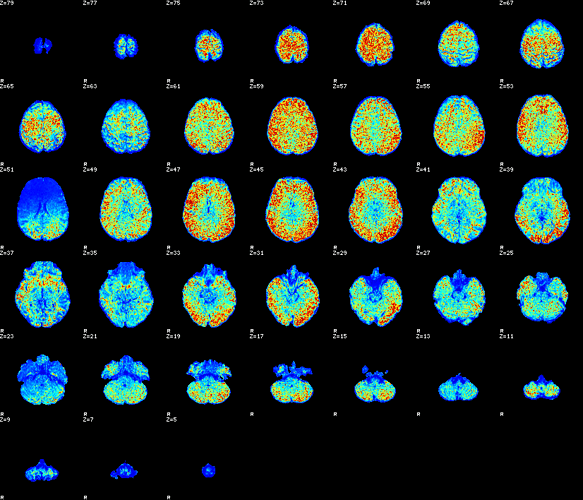
I collect mean/max values to find outliers at the group level. Sometimes outliers in these values can flag subjects with potential problems. Below is Eddy b=0 SNR image from one subject who was outlier in b=0 afni mean and max outlier values, for instance.
Those images are great! Do you have the code that generated these on GitHub somewhere so we can incorporate into dmriprep? I’ve also created an issue on our repo here: https://github.com/nipy/dmriprep/issues/13 to add your images to the QC report
At the moment they are part of a private lab repo on gitlab but I will pull out codes that concerns the dwi QC part to my github and share with you guys. I will let you know as soon as it’s done!
I’m also working on a pipeline for diffusion image processing https://github.com/pennbbl/qsiprep and across-software reconstruction. The preprocessing part is mostly done and the reconstruction part is coming along.
@mattcieslak: that looks great! Do I understand correctly that this pipeline will not work for HARDI- and DTI-oriented protocols? Looks like it might be complimentary to what we want to do with dmriprep. Maybe we could aim to merge streams eventually?
Thanks @Ariel_Rokem! The preprocessing part works with HARDI, just not single shell data. The reconstruction part works with any type of data that is appropriate for the reconstruction method. We’re actually planning on using dmriprep for preprocessing our single shell data and reconstructing the output with qsiprep reconstruction pipeline.
An update here - as of 0.4.1 qsiprep also works for DTI data. If the --hmc-model eddy option is specified, eddy is used. If an epi fieldmap is specified in BIDS for a scan, then TOPUP is also run and the fieldcoeffs are sent to eddy. We run CI tests on DTI input (https://github.com/PennBBL/qsiprep/blob/420ffbbc7e918b5e2354705a8b497fac0dd84bc1/.circleci/config.yml#L332) so I’m fairly sure this is working correctly. Definitely would welcome others to test it, though.
I think we should merge projects, using the preprocessing from qsiprep and the reports from dmriprep. We had discussed merging under the name dmriprep, but I know @oesteban is also working on a dmriprep and it would be potentially confusing to have two of them. I’m happy with either name, but would like to hear from everyone before renaming anything.