Thank you for looking into this. Would you have the possibility to run ANTs N4BiasFieldCorrection on your side (from a local installation of ANTS from example) on this problematic image? Just to understand if the problem is coming from the image itself or if it is a problem with the software execution within fmriprep.
Not sure that I know how to do that. The only installation available for me is if I shell into the singularity. But am kinda lost from that point onwards.
If you wanted to try from your singularity image, you could try a simple call like this:
singularity exec -B /PATH_TO_YOUR_FMRIPREP_WORKING_DIR:/out -B /PATH_TO_YOUR_TEST_OUTPUT:/work /PATH_TO_YOUR_SINGULARITY_IMAGES/fmriprep-23.0.2.simg N4BiasFieldCorrection -i /out/fmriprep_23_0_wf/single_subject_sid001429_wf/func_preproc_task_movie_run_07_wf/initial_boldref_wf/gen_avg/sub-sid001429_task-movie_run-07_bold_average.nii.gz -o /work/test_N4.nii.gz -v 1
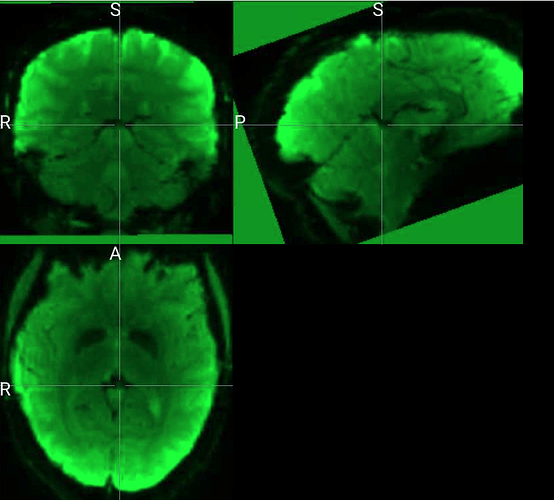
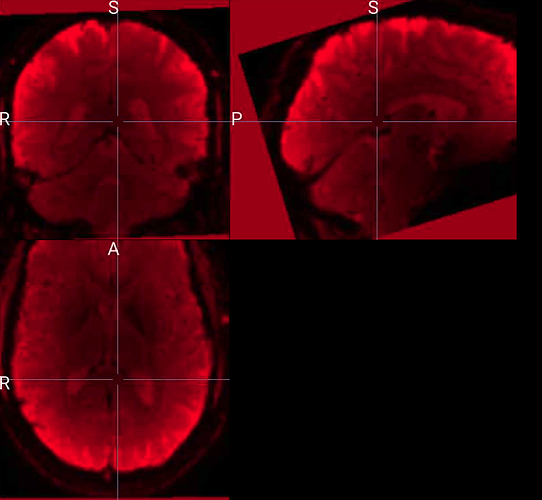
The input image for this subject doesn’t look noticeably different to the input for subjects for whom preprocessing completed successfully, at least as far as I can tell by visually inspecting in mricron. Attaching screenshots here in case helpful.
Can you try running N4BiasFieldCorrection on the image? If you look at @jsein’s last post, you can even run it in the container.
So I tried running the command as @jsein suggested and it ran fine and the output looks reasonable.
However, when i added the "-r --weight-image " argument that is in the fmriprep command it failed with the following error:
Running N4 for 3-dimensional images.
Mask not read. Using the entire image as the mask.
Original intensity range: [0, 515.159]
***********************************************************
Warning: Your input image contains nonpositive values
which could cause failure or problematic results. A
possible workaround would be to:
1. rescale your image to positive values e.g., [10,100].
2. run N4 on your rescaled image.
3. (optional) rescale the N4 output to the original
intensity range.
***********************************************************
Exception caught:
itk::ExceptionObject (0x21cfe60)
Location: "unknown"
File: /src/ants/build/staging/include/ITK-5.1/itkImageToImageFilter.hxx
Line: 220
Description: itk::ERROR: N4BiasFieldCorrectionImageFilter(0x21d24b0): Inputs do not occupy the same physical space!
InputImage Origin: [-1.2193752e+02, 9.1350460e+01, -8.8487409e+01], InputImageMaskImage Origin: [-1.2193241e+02, 9.1350059e+01, -8.8492116e+01]
Tolerance: 1.2000000e-05
InputImage Direction: 9.9962835e-01 2.7125358e-02 -2.7143980e-03
2.7125358e-02 -9.7980313e-01 1.9811622e-01
-2.7143979e-03 1.9811622e-01 9.8017478e-01
, InputImageMaskImage Direction: 9.9962782e-01 2.7228536e-02 -1.6832433e-03
2.7022163e-02 -9.7980314e-01 1.9813031e-01
-3.7455512e-03 1.9810203e-01 9.8017424e-01
Tolerance: 1.0000000e-06Ah, yeah. We’ve seen that recently. Upgrading to a more recent ANTs will fix it. Can you try testing on nipreps/fmriprep:unstable? (singularity exec ... docker://nipreps/fmriprep:unstable N4BiasFieldCorrection ... should work.)
Just to be clear. For that we will need to build the new image for the unstable version and then re-do the n4 command within the new container.
Yes. This is fixed in a new version of ANTs, but we don’t upgrade dependencies in Docker containers within a minor series. So the fixed version will be 23.1.0, which we hope to release very soon.
The N4 command runs through with the new container.
The question now is is it possible just ran those commands manually to have their outputs in the work folder and execute the rest of the pipeline with 23.0.2 to maintain consistency with the rest of the dataset.
If not, are there other changes coming up that will make participants processed with this version different?
I think this should work to make the problematic participant go through with fmriprep v 23.0.2.
I am glad you found the source of the bug!
Hi, could anyone please help me with running manually the N4 process via docker? (Not using singularity) so I can run the rest of my participants using 23.0.2 as well
I’m not sure how to go about it, i.e what command to use in order to run only the N4 process using the ‘unstable’ version and what command to use to run the rest of the pipeline only.
Thank you
Thank you very much
If you use fmriprep-docker, you can use your normal command, but add -i nipreps/fmriprep:unstable --shell to it. Then you’ll have a shell inside your container and access to all of the tools there.
It might be easier just to install ANTs locally, though: Release Eotapinoma · ANTsX/ANTs · GitHub
Thanks. Is the following command correct and will execute the regular workflow of fmriprep 23.0.2, except for the ‘N4BiasFieldCorrection’ for which it will use the unstable version?
Blockquote
docker run -it --rm ^
-v C:\Users\YossiY100\Desktop\2021_analysis\Nifti:/data:ro ^
-v C:\Users\YossiY100\Desktop\2021_analysis\derivatives:/out ^
-v C:\Users\YossiY100\Desktop\license.txt:/opt/freesurfer/license.txt:ro ^
-v C:\Users\YossiY100\Desktop\tmp:/mnt/c/Users/Yossi100/desktop/tmp ^
nipreps/fmriprep:23.0.2 ^
/data /out/fmriprep-23.0.2 ^
participant --participant-label sub-3453202111170831 ^
–fs-license-file /opt/freesurfer/license.txt ^
–ignore slicetiming ^
–low-mem ^
–mem-mb 12000 ^
–skip_bids_validation ^
–work-dir /mnt/c/Users/Yossi100/desktop/tmp ^
-i nipreps/fmriprep:unstable --shell N4BiasFieldCorrection
docker run -it --rm ^
-v C:\Users\YossiY100\Desktop\2021_analysis\Nifti:/data:ro ^
-v C:\Users\YossiY100\Desktop\2021_analysis\derivatives:/out ^
-v C:\Users\YossiY100\Desktop\license.txt:/opt/freesurfer/license.txt:ro ^
-v C:\Users\YossiY100\Desktop\tmp:/mnt/c/Users/Yossi100/desktop/tmp ^
--entrypoint bash nipreps/fmriprep:unstable
I’m Realizing that it is not clear to me if it is possible/how to execute the entire pipeline but only replace the N4BiasFieldCorrection commands with those from the new version and have the rest of the pipeline build around it.
I can manually create the output .nii.gz but i’m not sure how to get the entire pipeline to incorporate it. The commands above only seem to allow the manual creation part but not the reworking of the pipe around it. Unless i’m missing something
I understand what you mean.
I tried a couple of times running the command that failed using the “unstable” version, successfully manually creating the output .nii.gz but every time i let the 23.0.2 version run it tries running the N4 process, overrides that file and fails
Does anybody know of a way of stopping fmriprep from overriding the manually created N4Bias .nii.gz file, and instead keep it and incorporate it in the rest of the pipeline?
Hi, now I understand what you meant. I can install ANTs on the wsl. But could you please explain how could I update the ants version in the fmriprep 23.0.2 image so fmriprep will use the latest version of ants?
Thanks a lot
You would need to build the Docker image yourself. That’s beyond the scope of what I’m prepared to support.