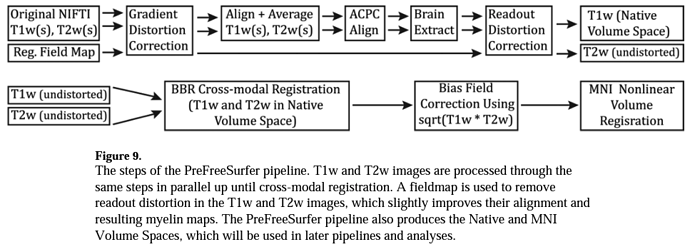
Hello Neurostars,
I’m processing the HCP-Aging 2.0 dataset with fMRIPrep and running into an issue. This issue is likely the result of a decision I made to use the output from HCP’s PreFreeSurferPipeline.sh as structural input for fMRIPrep.
I’ve made this decision because (1) I would like to do do all fMRI preprocessing using a pipeline that I’ve customized for a separate dataset, but (2) I would like to use the FreeSurfer output from HCP’s FreeSurfer pipeline because it has been thoroughly QCed. These files are very minimally processed; according to the code description:
# The primary purposes of the PreFreeSurfer Pipeline are:
#
# 1. To average any image repeats (i.e. multiple T1w or T2w images available)
# 2. To create a native, undistorted structural volume space for the subject
# * Subject images in this native space will be distortion corrected
# for gradient and b0 distortions and rigidly aligned to the axes
# of the MNI space. "Native, undistorted structural volume space"
# is sometimes shortened to the "subject's native space" or simply
# "native space".
# 3. To provide an initial robust brain extraction
# 4. To align the T1w and T2w structural images (register them to the native space)
# 5. To perform bias field correction
# 6. To register the subject's native space to the MNI space
I’ve symlinked the FreeSurfer input (i.e., $sub/mri/orig/{001,T2*}.mgz) to the subjects BIDS directory (i.e., $sub/ses-?/anat/$sub_ses-?_run-1_{T1,T2}w.nii.gz).
I’m not sure how to create a BIDS JSON file for these derivative images, so I used copies of a version of the images from an earlier HCP Pipeline (i.e., T1w_MPR_vNav_4e_e1e2_mean/${hcp_sub}_V1_MR_T1w_MPR_vNav_4e_e1e2_mean.json).
While running fmriprep using a containerized version of 22.1.0+0.gce344b39.dirty, I get the following error:
Command:
singularity run \
--cleanenv -B /projects \
/projects/p30952/environment/singularity_images/${FMRIPREP_VER}.simg \
${BIDS_DIR} \
${FMRIPREP_DIR} \
participant --participant-label ${subject} \
-w ${WORK_DIR} \
--skip_bids_validation \
--omp-nthreads 8 \
--nthreads 12 \
--mem_mb 24000 \
--fs-license-file ${FS_LICENSE} \
--fs-subjects-dir ${BIDS_DIR}/derivatives/freesurfer \
--output-spaces anat fsnative MNI152NLin2009cAsym fsLR fsaverage \
--cifti-output 91k \
--fd-spike-threshold 0.4 \
--dvars-spike-threshold 1.5 \
--dummy-scans 4
Error:
Node Name: fmriprep_22_1_wf.single_subject_HCA6002236_wf.anat_preproc_wf.anat_template_wf.t1w_ref_dimensions
File: /projects/b1145/bram/hcp-aging/derivatives/fmriprep-22.1.0/sub-HCA6002236/log/20230224-122507_f0b9a088-0c7d-4934-9bf2-1a88c2b34db5/crash-20230224-123908-brd2241-t1w_ref_dimensions-6fe94ef1-e8aa-4685-8de9-ef9333e2602e.txt
Working Directory: /projects/b1145/bram/work/fmriprep_22_1_wf/single_subject_HCA6002236_wf/anat_preproc_wf/anat_template_wf/t1w_ref_dimensions
Inputs:
max_scale: 3.0
t1w_list:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node t1w_ref_dimensions.
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/niworkflows/interfaces/images.py", line 436, in _run_interface
orig_imgs = np.vectorize(nb.load)(in_names)
File "/opt/conda/lib/python3.9/site-packages/numpy/lib/function_base.py", line 2304, in __call__
return self._vectorize_call(func=func, args=vargs)
File "/opt/conda/lib/python3.9/site-packages/numpy/lib/function_base.py", line 2382, in _vectorize_call
ufunc, otypes = self._get_ufunc_and_otypes(func=func, args=args)
File "/opt/conda/lib/python3.9/site-packages/numpy/lib/function_base.py", line 2342, in _get_ufunc_and_otypes
outputs = func(*inputs)
File "/opt/conda/lib/python3.9/site-packages/nibabel/loadsave.py", line 103, in load
matches, msg = _signature_matches_extension(filename, sniff)
File "/opt/conda/lib/python3.9/site-packages/nibabel/loadsave.py", line 65, in _signature_matches_extension
if sniff.startswith(expected_signature):
AttributeError: 'tuple' object has no attribute 'startswith'
My Questions:
- Is my logic in using FreeSurfer input for fMRIPrep flawed? If so, what files from HCP should I use?
- Is the solution as simple as creating more appropriate JSON files for the fMRIPrep input? How can I accomplish this for derivatives from a prior processing pipeline?
- I’m worried that the issue is within the header of the image files I’m using as structural inputs to fMRIPrep; should/how can I modify this?
Cheers,
Bram