Hi, I had fmriprep run on a single session dataset (1st time): a T1 and 6 functional scans (each the identical protocol/task). I’ve looked at this data with other tools before, it seemed pretty normal (meaning, I could plot the same post-registration voxel over six scans and see a similar retinotopy stimulus-evoked time course).
This was the command:
fmriprep-docker /Volumes/vision/Data/BIDS/bidsout /Volumes/vision/Data/new_pype/output participant --participant-label sub-01
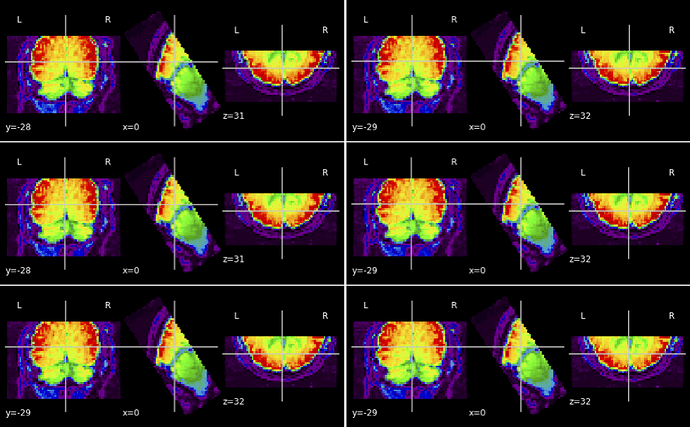
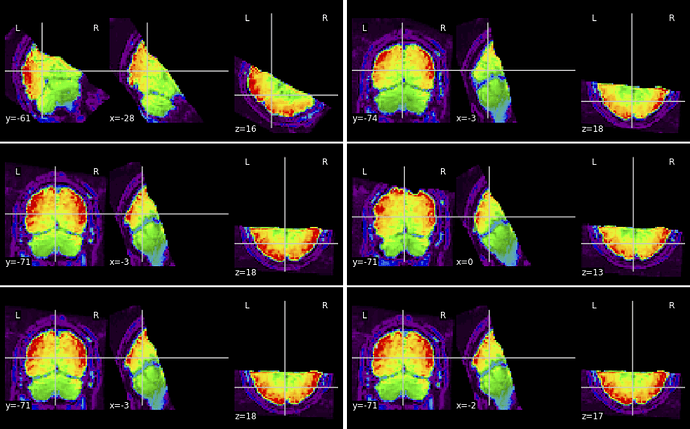
looking at the html summary output, I see that the first of 6 EPIs is extremely badly misaligned, like nonsense (turned sidewise, rotated 90deg, etc). On that scan I got the message " bbregister refinement rejected". It was rejected also on another scan, which is also poorly aligned but at least in the right universe.
Of the other 4 (bbregister succeeded), three look perfect but one is rather off.
How can I investigate where this went wrong? I used dcm2bids to convert the data this time, could it go wrong there (formatting problem?)?
thanks
-andrew
edit for more detail
What I had done previously that got nice results was a patched-together notebook from this nipype tutorial:
https://miykael.github.io/nipype_tutorial/notebooks/example_preprocessing.html
And the inputs to that were niftis that had been converted using the MATLAB dcm2nii function:
So I thought that since the underlying stuff was mostly the same, the dcm2bids → fmriprep would give similar results.