Summary of what happened:
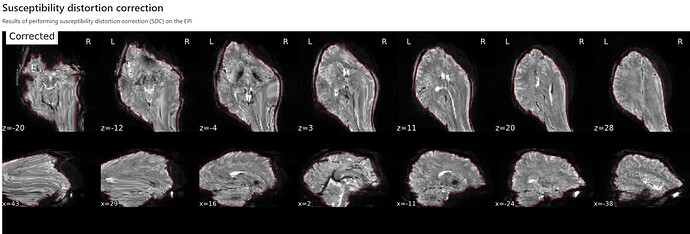
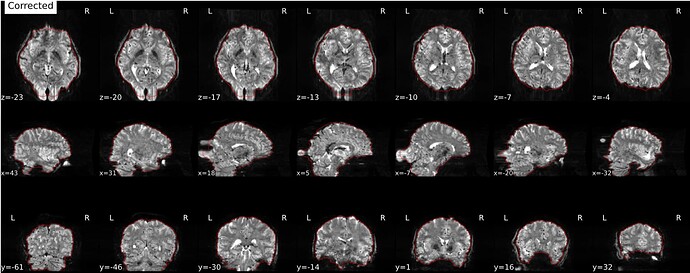
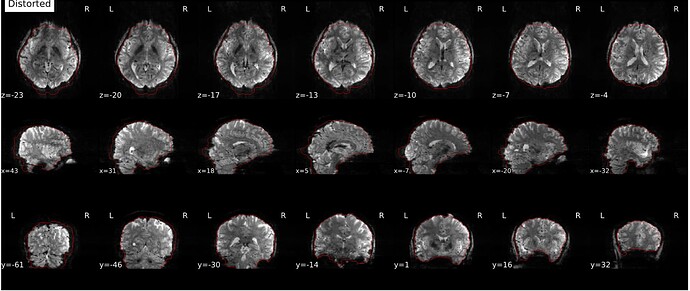
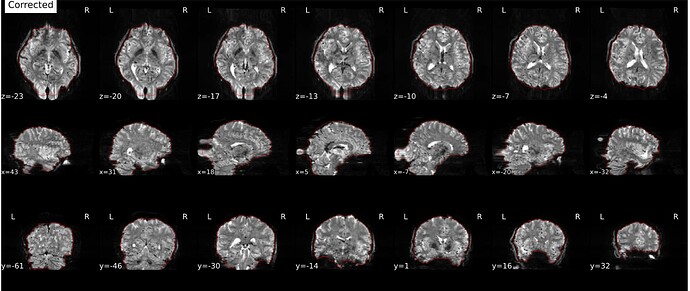
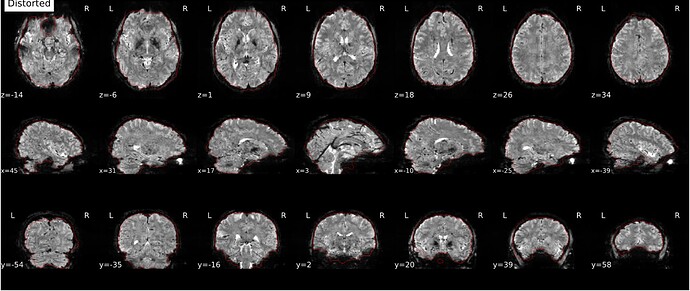
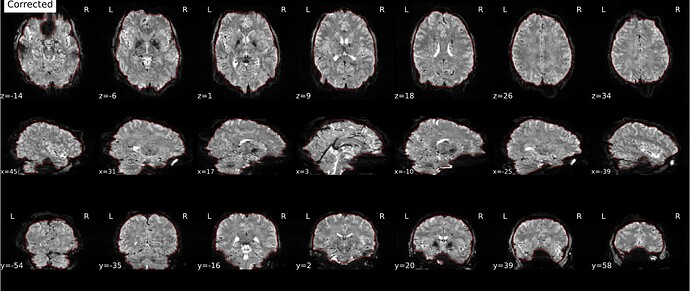
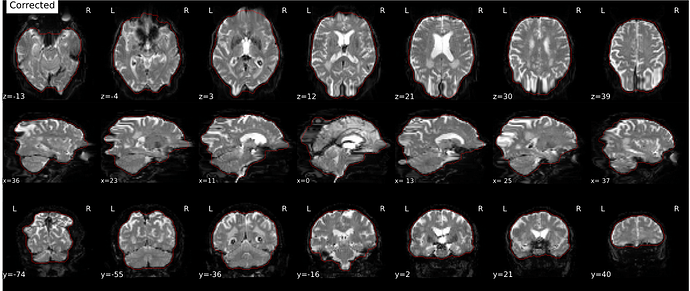
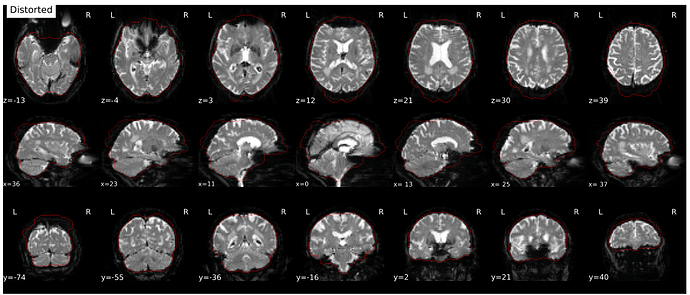
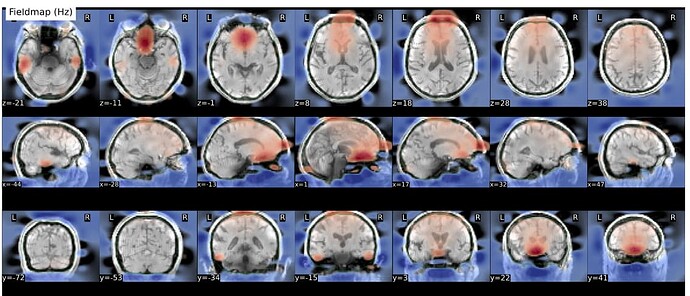
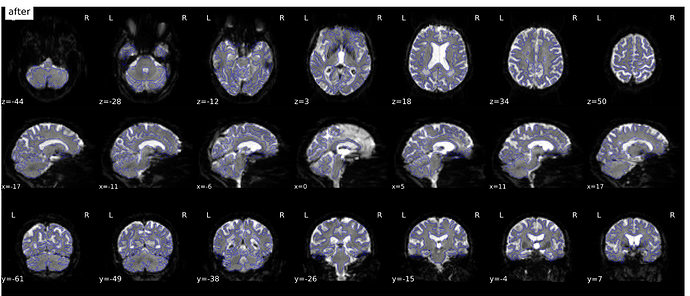
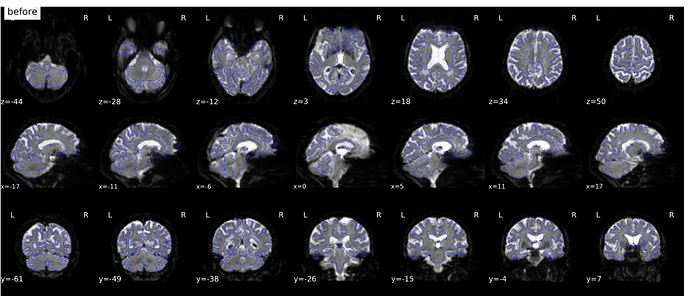
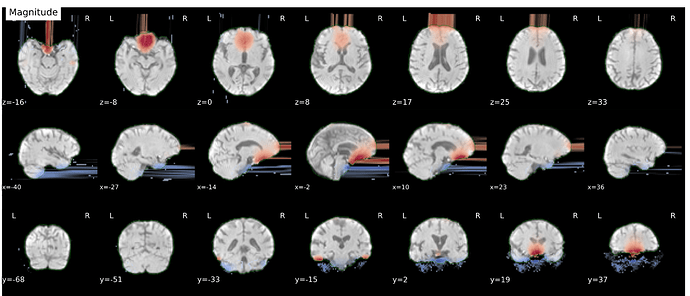
Hi everyone. We got some 7T fMRI data but encountered one issue during the preprocessing using fmriprep. As shown in the screenshots below, the SDC severely distorted the image. Two magnitude and one phase difference images were used for the correction. We tried the same preprocessing process on 5 subjects’ data, 4 out of 5 were distorted and there were errors raised for those subjects. We also tried the same process for 3T data and that works well.
As a fresher with fmriprep, I really have no idea what’s the issue and how to deal with it. If possible, I really hope to get some advice from people that is more experienced.
Thank you very much for any help in advance.
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep-docker \
--user $(id -u):$(id -g) \
bids_7T/ \
bids_7T/derivatives/ \
participant \
-w workdir/ \
--participant_label '+pid+' \
--ignore slicetiming \
--output-spaces T1w run func \
--fs-license-file /usr/local/freesurfer/license.txt \
--force-bbr \
--dummy-scans 10 \
--use-aroma \
--aroma-melodic-dimensionality -200 \
--return-all-components \
--fs-no-reconall \
--write-graph \
Version:
20.0.2
Environment (Docker, Singularity, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
I got one warning from the validator:
#### **Warning 1: [Code 102] TOO_FEW_AUTHORS**
[Click here for more information about this issue](https://neurostars.org/search?q=TOO_FEW_AUTHORS)
The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered based on the presence of only one author field. Please ignore if all contributors are already properly listed.
Relevant log outputs (up to 20 lines):
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node aroma_confounds.
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/utility/wrappers.py", line 142, in _run_interface
out = function_handle(**args)
File "", line 7, in _to_join
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/niworkflows/interfaces/utility.py", line 376, in _run_interface
raise ValueError("Number of columns in datasets do not match")
ValueError: Number of columns in datasets do not match