Hi all,
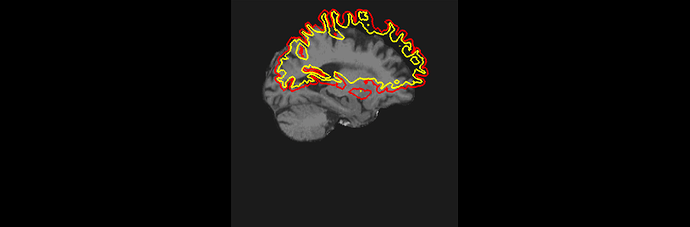
Even though in the “recon all.log”: it says: recon-all -s sub025 finished without errors, after checking the pial and white matter segmentations, very clear misplacement failures and part of dura mater are present (see attachment: segmentation misplacement and dura
I hope someone of you could help me with solving this error.
I am running recon all from the terminal of a linux machine: freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.0-2beb96c.
I tried recon all on a T1 of small vessel disease patients. I tried the command on several subjects and I got the same outcome: a bad allignment of pial and white surfaces.
For sake of curiosity, I tried the recon all command on another dataset (stroke) and with these data I got an hard failure.
recon all exited with errors:
ERROR: talairach_afd: Talairach Transform: transforms/talairach.xfm FAILED (p=0.0573, pval=0.0034 < threshold=0.0050). (see second recon all o35403143 with errors)
I would like to add the recon all log to this post but apparently new users are not allowed to do so.
Any advice is appreciated!
Thank you in advance!
cheers
Ileana