This is correct. ANTs produces ITK transforms as .mat files for affines and .nii.gz for warps, or .h5 when you have a chain of transforms (which is often the case). For SDC, there is no shift or rotation, just a warp.
I’m going to get a bit in the weeds here, but it may be useful to have this written down for reference. In recent fMRIPrep (I’ll need to check which versions), we actually perform SDC in two ways. One for the BOLD native space (which is relevant to the multi-echo case), and one for the T1w/MNI space.
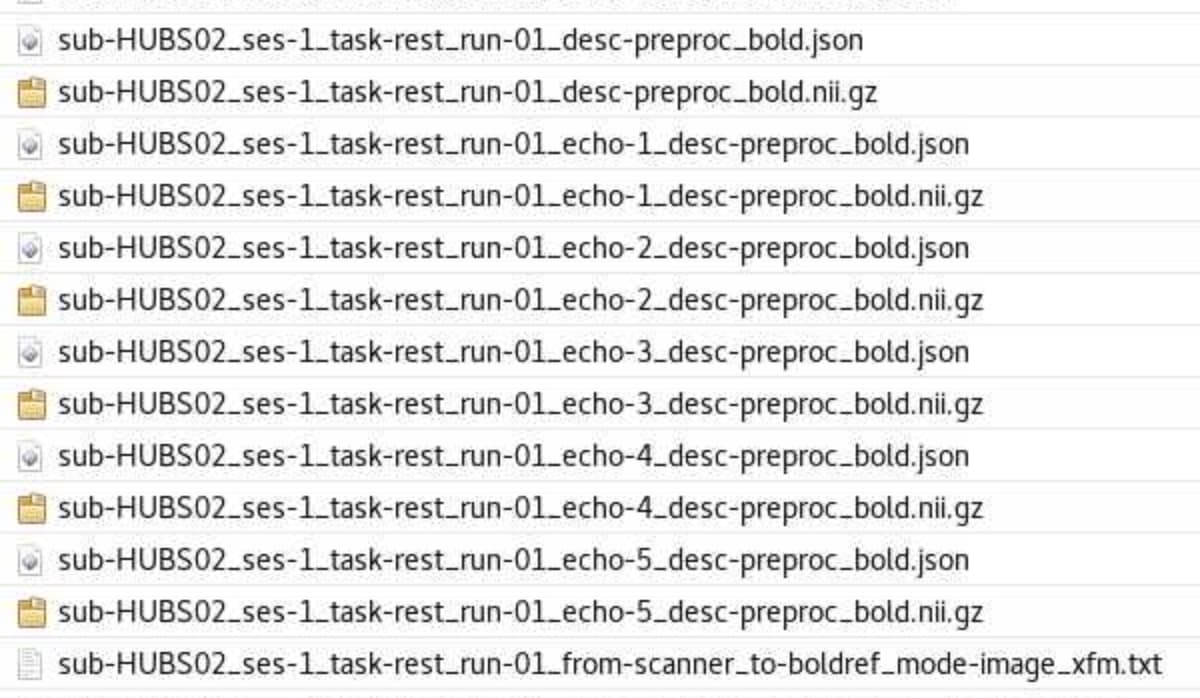
Here’s an example for the BOLD native one from our test multi-echo dataset:
$ ls fmriprep_23_1_wf/single_subject_02_wf/func_preproc_task_cuedSGT_run_01_echo_1_wf/unwarp_wf/_echoidx_0/resample/
00000 _node.pklz vol0001_unwarped.nii.gz vol0003_xfm.nii.gz
00001 _report/ vol0001_xfm.nii.gz vol0004_field.nii.gz
00002 result_resample.pklz vol0002_field.nii.gz vol0004_unwarped.nii.gz
00003 vol0000_field.nii.gz vol0002_unwarped.nii.gz vol0004_xfm.nii.gz
00004 vol0000_unwarped.nii.gz vol0002_xfm.nii.gz
_0x4113a609562a41f17c930c47cec3b759.json vol0000_xfm.nii.gz vol0003_field.nii.gz
_inputs.pklz vol0001_field.nii.gz vol0003_unwarped.nii.gz
What you’re seeing here is:
00000 - Head motion transform in ITK format. (Bad name because it’s not used anywhere else.)vol00000_field.nii.gz - The B0 fieldmap resampled into vol00000 using the HMC transformvol00000_xfm.nii.gz - The ITK format deformation field for volume 00000, derived from the field.vol00000_unwarped.nii.gz - The corrected vol00000
(Since only *_unwarped.nii.gz are used by later nodes, the others will generally be deleted unless you have nipype settings configured to avoid deleting unused outputs, which I do for debugging.)
This is an early version of a technique we’ve been developing to reduce the interactions of susceptibility effects and head motion. Basically, the inhomogeneities that cause the distortion happen based on the location of structures in the head, but the distortion occurs in one direction relative to the scanner bore. When you move relative to that direction vector, the distortion changes direction, so can’t be fully corrected by performing head motion correction and then applying a warp as if the head was motionless.
That said, this approach is wildly expensive and gets more complicated when you add additional transforms. So we hadn’t implemented it for other spaces yet, keeping the resampling we’ve been doing for years. For small motion, the effect should be small.
Now, it turns out it does not need to be wildly expensive to do the correction correctly. The 23.2 release will resample all series in the same way, and it will be much cheaper. If you really want to dive into it, I’ve written up the theory here: https://hackmd.io/@effigies/rethinking-resampling