Dear all,
I have a question about spatial normalization and resample in stroke patients. I would greatly appreciate the help, and I apologize if it is a silly question!
I need the input lesion segmentation files have 1mm3 voxel dimensions and images dimensions of 182x218x182.
However, I have some obstacles that I have not been able to resolve yet.
-
It would be essential if I could use the Clinical Toolbox (by SPM12) because I need to normalize MRI and CT images. However, as little as I know so far (I may be wrong), it does not have that dimension of the MNI space. Am I right?
-
The SPM12’s template is of “average brain size”, as well as SMP12 uses an “own space” based on scans acquired within the IXI data set (IXI549Space). So, first of all, I cannot consider that the clinical toolbox (older people template), will work in a program that asks for lesion files registered in the MNI space, right? In addition, if I changed the dimension of the lesion mask that was normalized by the Clinical Toolbox, using the “resample” function, having as a “reference image” an MNI template with a dimension of 182x218x182. It’s wrong? Or, does the “resample” have to be done before spatial normalization?
Thanks so much,
Luan.
Andrew Jahn shows how to make the “resample” in SPM12: https://www.youtube.com/watch?v=rvW-D5o3ALA
Rorden, C., Bonilha, L., Fridriksson, J.,et al.(2012). Age-specific CT and MRI templates for spatial normalization. Neuroimage, 61, 957–965. https://doi.org/10.1016/j.neuroimage.2012.03.020
-
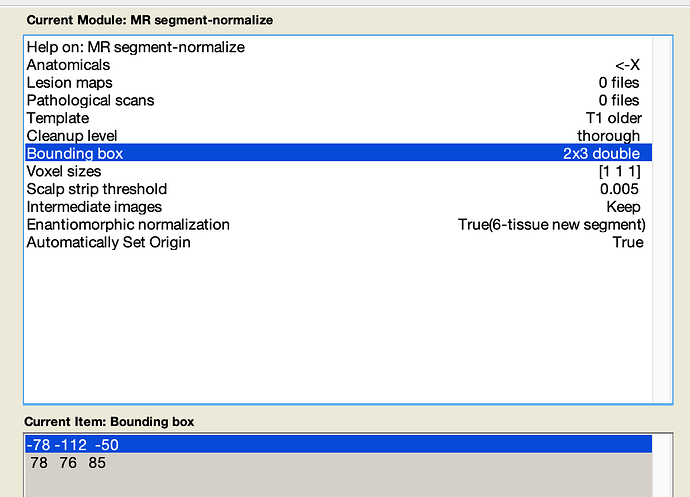
You can adjust the “bounding box” in the Clinical toolbox (and other SPM normalization scripts) to adjust the field-of-view of the resampled data, and you can adjust the voxel size to adjust the resolution. The initial values are the SPM defaults. As I recall, SPM images are typically replaced to an odd number of voxels in the left-right dimension (e.g. 181 vs 182), so the axis of symmetry runs through the center of voxels. You may want to specify why the 182x218x182 is specific for your application, rather than simply making images from all individuals consistent.
-
Yes, SPM normalizes brains to have be the size of an average brain, while the MNI template is larger than the average brain (as described in the original paper on the MNI template). Therefore, you really do not want to mix-and-match images warped to an MNI template with ANTS, FSL, etc. and from SPM. Further, note that the Clinical toolbox allows you to select between the “T1 older” and the “T1 younger” templates. The longer template matches the SPM defaults. The older template is based on neurologically healthy individuals age-matched to stroke patients. The older template demonstrates cerebral atrophy as one would expect. The intention is to maximize the match and minimize distortions. However, you should not mix and match participants aligned to different templates. The older template does show considerably larger ventricles, wider sulci, and the boundary of the cortex is clearly a bit different near the frontal cortex.
When working with clinical scans, please carefully visually inspect each solution. Most routines, including the clinical toolbox have issues when confronted with wide diploid space, which is associated with stroke risk factors. At some stage, I will release a new script for enantiomorphic normalization of stroke brains that leverages CAT12. In my experience, this provides the most robust solution to this problem, outperforming any solutions I have seen, including those I developed myself specifically for this problem.
1 Like
Dear Rorder,
Initially, I would like to thank you for your time and immense help! Your answer was beneficial to me! Also, I would like to apologize for the delay in my response. The current scenario is making my activities very difficult.
Best,
Luan
You may also want to look here for a tool that seems to use enantiomorphic normalization with ANTS. Since ANTS uses the MNI template rather than an average brain, it will match that shape and size.
You could also use this function to create enantiomrophically healed brains, and then use any normalization function you want: you use the ‘healed’ brain to compute the normalization, and then you use the warping transform on the original lesioned brains (FSL calls this a ‘shadow registration’ where the registration ins based on a different image than what it is applied to).
1 Like
Dear Rorden,
Many thanks for the reply! I’m learning a lot!
The “Normalisation” tool from the BCBlab toolkit is handy. It provides a T1 registration for the MNI template in the desired dimensions (182x218x182). However, I would like to use both MRI and CT images of stroke patients in my work. That was one of the reasons I wanted to use Clinical Toolbox. Do you have any suggestions on this topic (CT template)?
Regarding the second suggestion, could you tell me where I can see more about this “shadow registration”? I was not successful in finding more information on this topic on the FSL website.
Best,
Luan
Hi Luan
I would like to try the BCBlab toolkit to try the T1 registration for the MNI template in the dimensions you mentioned (182x218x182).
Would you be willing to share how the install and open the BCBLab toolkit. I am using Linux Ubuntu 20.04. I was able to download the toolkit, but am not able to open it. Do you have any tips?
Thank you
Marie