Summary of what happened:
Dear fMRIprep experts,
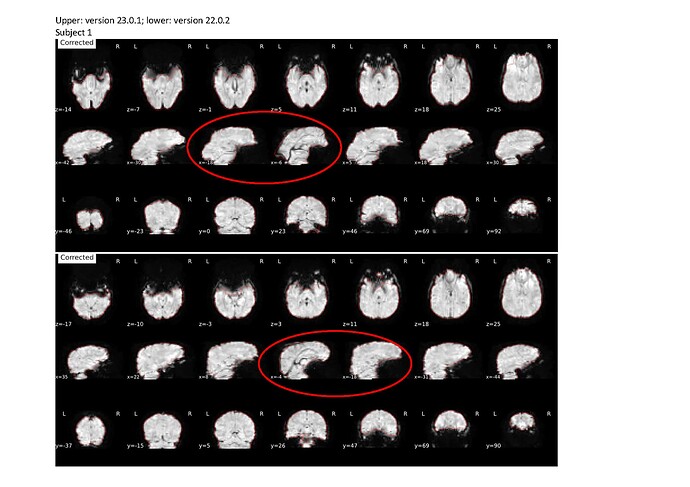
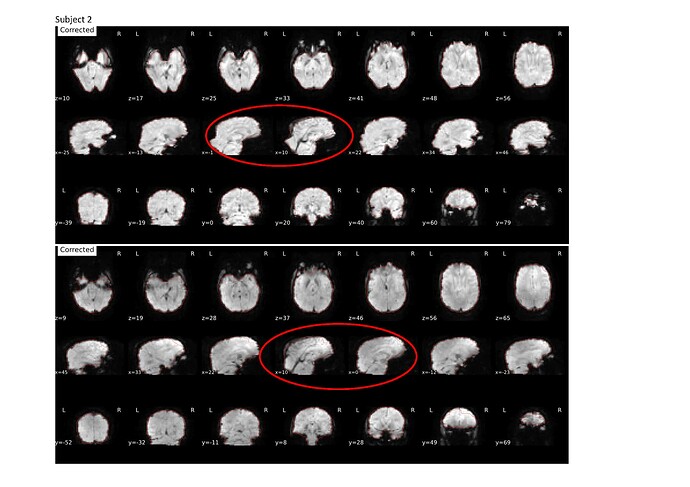
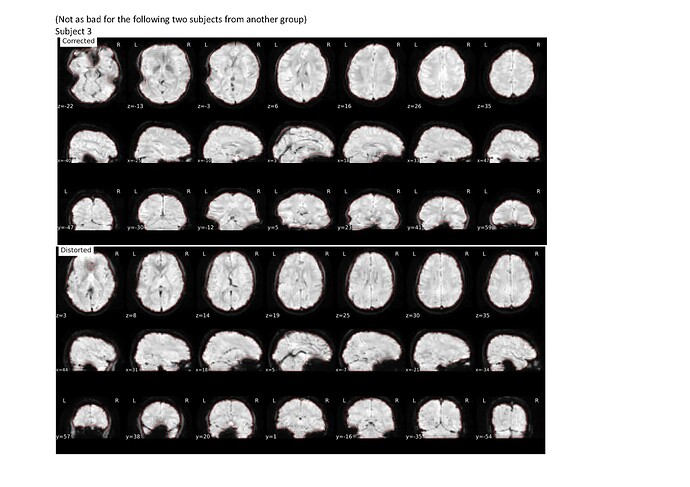
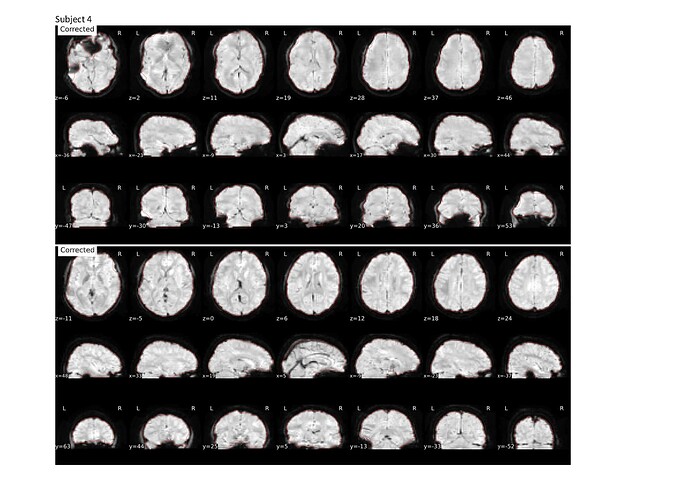
I previously used the --use-syn-sdc for two sets of data using version 22.0.2 then re-ran the pre-processing after deleting all the output files using the latest version 23.0.1 and noticed that the images are more distorted compared to that using the older version (see attached screenshots below; subject 1 and 2 are from a different group than subject 3 and 4).
Not sure if this is relevant but I previously did not include the --cifti-output flag when using version 22.0.2.
Any guidance on what the cause of this discrepancy might be and if I should stick to version 22.0.2 or another slightly older version will be greatly appreciated!
Command used (and if a helper script was used, a link to the helper script or the command generated):
export SINGULARITYENV_TEMPLATEFLOW_HOME="${myproject}/templateflow"
singularity run -B "${myproject}":"${myproject}" --cleanenv "${myproject}/fmriprep-23.0.1.simg" --skip-bids-validation "${myproject}/MA_1_BIDS" "${myproject}/MA1_SDC_Processed" --mem_mb 40000 participant --participant-label "${SUBJECT}" --fs-license-file "${myproject}/license.txt" --output-spaces MNI152NLin6Asym:res-2 MNI152NLin2009cAsym:res-2 --use-aroma --use-syn-sdc --cifti-output -w /tmp/
Version:
fmriprep-23.0.1.simg
Environment (Docker, Singularity, custom installation):
Singularity
Relevant log outputs (up to 20 lines):
Output files for subject 1 and 2 are attached.
output_subject_1.txt (858.8 KB)
output_subject_2.txt (856.6 KB)
output_subject3.txt (901.7 KB)
output_subject4.txt (901.8 KB)