Summary of what happened:
Hi all,
We’re trying to put some intensively sampled data through fmriprep, 4 subjects ~ 10 sessions each on our HPC. The data are multi-echo with 4 or 5 runs of functional data, depending on the session. Like in some other posts I’ve read, we’re trying to avoid using the longitudinal flag and instead process each session separately. To do that, I’ve created a script that generates a subject-session-specific bids config file and fmriprep command that I can submit one-by-one. I’ve also created session-specific derivative and scratch directories to avoid issues there.
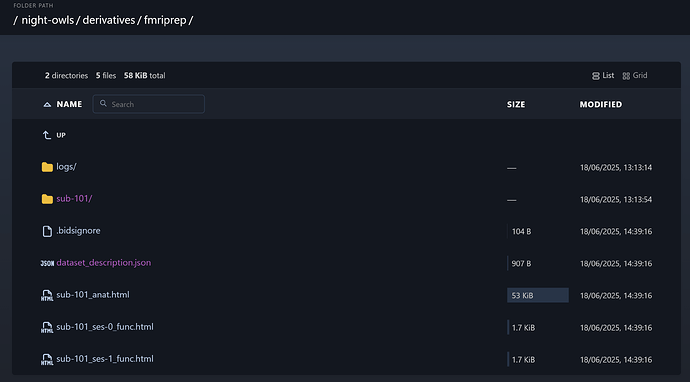
It looks like things mostly worked, but something weird is going on with the summary .html outputs that is breaking ses-0x into one _ses-0_func.html file and another ses-x_func.html file (see screenshot below). Both of these files are 2KB and have nothing when I open them (presumably because no outputs match ses-0 or ses-x?).
As far as we can tell so far, the rest of the outputs look correct. fmriprep indeed said it ran succesfully. If this is just a syntax error for the output summary, I suppose we can move forward, but I would like to confirm nothing deeper is going on here.
I’ve now replicated this with ses-01 twice as well as ses-02 for our first subject.
Command used (and if a helper script was used, a link to the helper script or the command generated):
This script generates our bids config files and fmriprep commands:
#!/bin/bash
# ensure paths are correct
projectname=night-owls
maindir=/gpfs/scratch/tug87422/smithlab-shared/$projectname
scriptdir=$maindir/code
bidsdir=$maindir/bids
logdir=$maindir/logs
mkdir -p $logdir
#Keep scratch and derivatives dirs separate by sessions
for ses in {01..12}; do
if [ ! -d "$maindir/scratch/ses-$ses" ]; then
mkdir -p "$maindir/scratch/ses-$ses"
fi
if [ ! -d "$maindir/derivatives/ses-$ses" ]; then
mkdir -p "$maindir/derivatives/ses-$ses"
fi
done
# Generate configs and PBS scripts
subjects=(sub-101)
for sub in ${subjects[@]}; do
## for sessions that have a functional image..
sessions=( $(
find "$bidsdir/$sub" \
-type f \
-path "*/ses-[0-9][0-9]/func/*.nii.gz" \
2>/dev/null \
| sed -n 's|.*/\(ses-[0-9][0-9]\)/.*|\1|p' \
| sort -u
) )
for ses in "${sessions[@]}"; do
sesid=${ses#ses-}
## Create subject/session-specific BIDS filter file
configfile=$maindir/code/fmriprep/fmriprep_config_${sub}_${ses}.json
cat <<EOF > "$configfile"
{
"t1w": {"datatype": "anat", "suffix": "T1w", "session": "$sesid"},
"sbref": {"datatype": "func", "suffix": "sbref", "part": [null, "mag"], "session": "$sesid"},
"bold": {"datatype": "func", "suffix": "bold", "part": [null, "mag"], "session": "$sesid"}
}
EOF
# Generate subject/session-specific PBS script
scriptfile=$maindir/code/fmriprep/fmriprep_${sub}_${ses}.sh
cat <<EOF > "$scriptfile"
#!/bin/bash
#PBS -l walltime=8:00:00
#PBS -N fmriprep-${sub}-${ses}
#PBS -q normal
#PBS -l nodes=1:ppn=14
# load modules and go to workdir
module load fsl/6.0.2
source \$FSLDIR/etc/fslconf/fsl.sh
module load singularity
cd \$PBS_O_WORKDIR
# ensure paths are correct
projectname=night-owls
maindir=/gpfs/scratch/tug87422/smithlab-shared/\$projectname
scriptdir=\$maindir/code
bidsdir=\$maindir/bids
logdir=\$maindir/logs
mkdir -p \$logdir
TEMPLATEFLOW_DIR=/gpfs/scratch/tug87422/smithlab-shared/tools/templateflow
MPLCONFIGDIR_DIR=/gpfs/scratch/tug87422/smithlab-shared/tools/mplconfigdir
export SINGULARITYENV_TEMPLATEFLOW_HOME=/opt/templateflow
export SINGULARITYENV_MPLCONFIGDIR=/opt/mplconfigdir
singularity run --cleanenv \\
-B \${TEMPLATEFLOW_DIR}:/opt/templateflow \\
-B \${MPLCONFIGDIR_DIR}:/opt/mplconfigdir \\
-B \$maindir:/base \\
-B /gpfs/scratch/tug87422/smithlab-shared/tools/licenses:/opts \\
-B \$maindir/scratch/$ses:/scratch \\
/gpfs/scratch/tug87422/smithlab-shared/tools/fmriprep-24.1.1.simg \\
/base/bids /base/derivatives/$ses \\
participant --participant_label $sub \\
--stop-on-first-crash \\
--skip-bids-validation \\
--nthreads 14 \\
--me-output-echos \\
--output-spaces MNI152NLin6Asym \\
--bids-filter-file /base/code/fmriprep/fmriprep_config_${sub}_${ses}.json \\
--fs-no-reconall --fs-license-file /opts/fs_license.txt \\
-w /scratch
EOF
chmod +x "$scriptfile"
done
done
Here is an example of what the config file and fmriprep script look like for sub-101, which was submitted as qsub fmriprep_sub-101_ses-01.sh
Config:
{
"t1w": {"datatype": "anat", "suffix": "T1w", "session": "01"},
"sbref": {"datatype": "func", "suffix": "sbref", "part": [null, "mag"], "session": "01"},
"bold": {"datatype": "func", "suffix": "bold", "part": [null, "mag"], "session": "01"}
}
Call:
#PBS -l walltime=8:00:00
#PBS -N fmriprep-sub-101-ses-01
#PBS -q normal
#PBS -l nodes=1:ppn=14
# load modules and go to workdir
module load fsl/6.0.2
source $FSLDIR/etc/fslconf/fsl.sh
module load singularity
cd $PBS_O_WORKDIR
# ensure paths are correct
projectname=night-owls
maindir=/gpfs/scratch/tug87422/smithlab-shared/$projectname
scriptdir=$maindir/code
bidsdir=$maindir/bids
logdir=$maindir/logs
mkdir -p $logdir
TEMPLATEFLOW_DIR=/gpfs/scratch/tug87422/smithlab-shared/tools/templateflow
MPLCONFIGDIR_DIR=/gpfs/scratch/tug87422/smithlab-shared/tools/mplconfigdir
export SINGULARITYENV_TEMPLATEFLOW_HOME=/opt/templateflow
export SINGULARITYENV_MPLCONFIGDIR=/opt/mplconfigdir
singularity run --cleanenv \
-B ${TEMPLATEFLOW_DIR}:/opt/templateflow \
-B ${MPLCONFIGDIR_DIR}:/opt/mplconfigdir \
-B $maindir:/base \
-B /gpfs/scratch/tug87422/smithlab-shared/tools/licenses:/opts \
-B $maindir/scratch/ses-01:/scratch \
/gpfs/scratch/tug87422/smithlab-shared/tools/fmriprep-24.1.1.simg \
/base/bids /base/derivatives/ses-01 \
participant --participant_label sub-101 \
--stop-on-first-crash \
--skip-bids-validation \
--nthreads 14 \
--me-output-echos \
--output-spaces MNI152NLin6Asym \
--bids-filter-file /base/code/fmriprep/fmriprep_config_sub-101_ses-01.json \
--fs-no-reconall --fs-license-file /opts/fs_license.txt \
-w /scratch
Version:
24.1.1
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
Here is the /func output, which looks normal as far as we can tell.
sub-101_ses-01_task-mid_run-1_echo-1_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-1_echo-1_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-1_echo-2_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-1_echo-2_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-1_echo-3_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-1_echo-3_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-1_echo-4_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-1_echo-4_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-1_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
sub-101_ses-01_task-mid_run-1_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
sub-101_ses-01_task-mid_run-1_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
sub-101_ses-01_task-mid_run-1_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
sub-101_ses-01_task-mid_run-1_part-mag_desc-brain_mask.json
sub-101_ses-01_task-mid_run-1_part-mag_desc-brain_mask.nii.gz
sub-101_ses-01_task-mid_run-1_part-mag_desc-confounds_timeseries.json
sub-101_ses-01_task-mid_run-1_part-mag_desc-confounds_timeseries.tsv
sub-101_ses-01_task-mid_run-1_part-mag_desc-coreg_boldref.json
sub-101_ses-01_task-mid_run-1_part-mag_desc-coreg_boldref.nii.gz
sub-101_ses-01_task-mid_run-1_part-mag_desc-hmc_boldref.json
sub-101_ses-01_task-mid_run-1_part-mag_desc-hmc_boldref.nii.gz
sub-101_ses-01_task-mid_run-1_part-mag_space-MNI152NLin6Asym_boldref.json
sub-101_ses-01_task-mid_run-1_part-mag_space-MNI152NLin6Asym_boldref.nii.gz
sub-101_ses-01_task-mid_run-1_part-mag_space-MNI152NLin6Asym_desc-brain_mask.json
sub-101_ses-01_task-mid_run-1_part-mag_space-MNI152NLin6Asym_desc-brain_mask.nii.gz
sub-101_ses-01_task-mid_run-1_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-1_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-1_space-MNI152NLin6Asym_T2starmap.json
sub-101_ses-01_task-mid_run-1_space-MNI152NLin6Asym_T2starmap.nii.gz
sub-101_ses-01_task-mid_run-2_echo-1_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-2_echo-1_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-2_echo-2_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-2_echo-2_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-2_echo-3_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-2_echo-3_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-2_echo-4_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-2_echo-4_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-2_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
sub-101_ses-01_task-mid_run-2_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
sub-101_ses-01_task-mid_run-2_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
sub-101_ses-01_task-mid_run-2_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
sub-101_ses-01_task-mid_run-2_part-mag_desc-brain_mask.json
sub-101_ses-01_task-mid_run-2_part-mag_desc-brain_mask.nii.gz
sub-101_ses-01_task-mid_run-2_part-mag_desc-confounds_timeseries.json
sub-101_ses-01_task-mid_run-2_part-mag_desc-confounds_timeseries.tsv
sub-101_ses-01_task-mid_run-2_part-mag_desc-coreg_boldref.json
sub-101_ses-01_task-mid_run-2_part-mag_desc-coreg_boldref.nii.gz
sub-101_ses-01_task-mid_run-2_part-mag_desc-hmc_boldref.json
sub-101_ses-01_task-mid_run-2_part-mag_desc-hmc_boldref.nii.gz
sub-101_ses-01_task-mid_run-2_part-mag_space-MNI152NLin6Asym_boldref.json
sub-101_ses-01_task-mid_run-2_part-mag_space-MNI152NLin6Asym_boldref.nii.gz
sub-101_ses-01_task-mid_run-2_part-mag_space-MNI152NLin6Asym_desc-brain_mask.json
sub-101_ses-01_task-mid_run-2_part-mag_space-MNI152NLin6Asym_desc-brain_mask.nii.gz
sub-101_ses-01_task-mid_run-2_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.json
sub-101_ses-01_task-mid_run-2_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz
sub-101_ses-01_task-mid_run-2_space-MNI152NLin6Asym_T2starmap.json
sub-101_ses-01_task-mid_run-2_space-MNI152NLin6Asym_T2starmap.nii.gz
sub-101_ses-01_task-rest_run-1_echo-1_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-rest_run-1_echo-1_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-rest_run-1_echo-2_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-rest_run-1_echo-2_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-rest_run-1_echo-3_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-rest_run-1_echo-3_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-rest_run-1_echo-4_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-rest_run-1_echo-4_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-rest_run-1_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
sub-101_ses-01_task-rest_run-1_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
sub-101_ses-01_task-rest_run-1_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
sub-101_ses-01_task-rest_run-1_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
sub-101_ses-01_task-rest_run-1_part-mag_desc-brain_mask.json
sub-101_ses-01_task-rest_run-1_part-mag_desc-brain_mask.nii.gz
sub-101_ses-01_task-rest_run-1_part-mag_desc-confounds_timeseries.json
sub-101_ses-01_task-rest_run-1_part-mag_desc-confounds_timeseries.tsv
sub-101_ses-01_task-rest_run-1_part-mag_desc-coreg_boldref.json
sub-101_ses-01_task-rest_run-1_part-mag_desc-coreg_boldref.nii.gz
sub-101_ses-01_task-rest_run-1_part-mag_desc-hmc_boldref.json
sub-101_ses-01_task-rest_run-1_part-mag_desc-hmc_boldref.nii.gz
sub-101_ses-01_task-rest_run-1_part-mag_space-MNI152NLin6Asym_boldref.json
sub-101_ses-01_task-rest_run-1_part-mag_space-MNI152NLin6Asym_boldref.nii.gz
sub-101_ses-01_task-rest_run-1_part-mag_space-MNI152NLin6Asym_desc-brain_mask.json
sub-101_ses-01_task-rest_run-1_part-mag_space-MNI152NLin6Asym_desc-brain_mask.nii.gz
sub-101_ses-01_task-rest_run-1_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.json
sub-101_ses-01_task-rest_run-1_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz
sub-101_ses-01_task-rest_run-1_space-MNI152NLin6Asym_T2starmap.json
sub-101_ses-01_task-rest_run-1_space-MNI152NLin6Asym_T2starmap.nii.gz
sub-101_ses-01_task-sharedreward_run-1_echo-1_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-1_echo-1_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-1_echo-2_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-1_echo-2_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-1_echo-3_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-1_echo-3_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-1_echo-4_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-1_echo-4_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-1_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
sub-101_ses-01_task-sharedreward_run-1_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
sub-101_ses-01_task-sharedreward_run-1_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
sub-101_ses-01_task-sharedreward_run-1_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-brain_mask.json
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-brain_mask.nii.gz
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-confounds_timeseries.json
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-confounds_timeseries.tsv
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-coreg_boldref.json
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-coreg_boldref.nii.gz
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-hmc_boldref.json
sub-101_ses-01_task-sharedreward_run-1_part-mag_desc-hmc_boldref.nii.gz
sub-101_ses-01_task-sharedreward_run-1_part-mag_space-MNI152NLin6Asym_boldref.json
sub-101_ses-01_task-sharedreward_run-1_part-mag_space-MNI152NLin6Asym_boldref.nii.gz
sub-101_ses-01_task-sharedreward_run-1_part-mag_space-MNI152NLin6Asym_desc-brain_mask.json
sub-101_ses-01_task-sharedreward_run-1_part-mag_space-MNI152NLin6Asym_desc-brain_mask.nii.gz
sub-101_ses-01_task-sharedreward_run-1_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-1_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-1_space-MNI152NLin6Asym_T2starmap.json
sub-101_ses-01_task-sharedreward_run-1_space-MNI152NLin6Asym_T2starmap.nii.gz
sub-101_ses-01_task-sharedreward_run-2_echo-1_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-2_echo-1_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-2_echo-2_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-2_echo-2_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-2_echo-3_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-2_echo-3_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-2_echo-4_part-mag_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-2_echo-4_part-mag_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-2_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
sub-101_ses-01_task-sharedreward_run-2_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
sub-101_ses-01_task-sharedreward_run-2_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
sub-101_ses-01_task-sharedreward_run-2_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-brain_mask.json
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-brain_mask.nii.gz
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-confounds_timeseries.json
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-confounds_timeseries.tsv
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-coreg_boldref.json
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-coreg_boldref.nii.gz
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-hmc_boldref.json
sub-101_ses-01_task-sharedreward_run-2_part-mag_desc-hmc_boldref.nii.gz
sub-101_ses-01_task-sharedreward_run-2_part-mag_space-MNI152NLin6Asym_boldref.json
sub-101_ses-01_task-sharedreward_run-2_part-mag_space-MNI152NLin6Asym_boldref.nii.gz
sub-101_ses-01_task-sharedreward_run-2_part-mag_space-MNI152NLin6Asym_desc-brain_mask.json
sub-101_ses-01_task-sharedreward_run-2_part-mag_space-MNI152NLin6Asym_desc-brain_mask.nii.gz
sub-101_ses-01_task-sharedreward_run-2_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.json
sub-101_ses-01_task-sharedreward_run-2_part-mag_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz
sub-101_ses-01_task-sharedreward_run-2_space-MNI152NLin6Asym_T2starmap.json
sub-101_ses-01_task-sharedreward_run-2_space-MNI152NLin6Asym_T2starmap.nii.gz