Summary of what happened:
I’m trying to use XCP-D to calculate the functional connectivity matrices from data preprocessed by fmriprep. However, some ROIs show n/a in whole timeseries consistently across different subjects. It may be worth noting that one subject has 7 fmri (3 with specific tasks while other 4 corresponding to two runs of resting-state with two directions). And resting-state image in dir-AP shows better result with no n/a value in most cases.
Command used (and if a helper script was used, a link to the helper script or the command generated):
To save the space, I post the main arguments of XCP-D and fmriprep:
XCP-D:
--participant-label ${1} \
--fs-license-file ${fs_dir} \
-w ${work_dir} \
-v \
--dcan-qc \
--nthreads 8 \
--omp-nthreads 2
fmriprep:
--participant-label ${1} \
--fs-license-file ${fs_dir} \
-w ${work_dir} \
-vv \
--nprocs 16 \
--omp-nthreads 8
Both of them actually are almost in default settings.
Version:
XCP-D: 0.5.2
fmriprep: 23.1.4
Environment (Docker, Singularity, custom installation):
Singularity
Screenshots / relevant information:
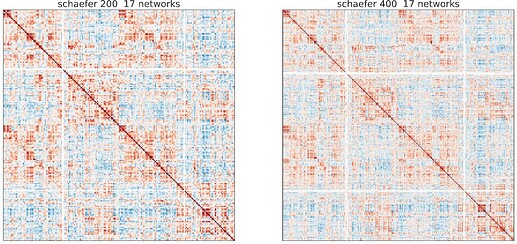
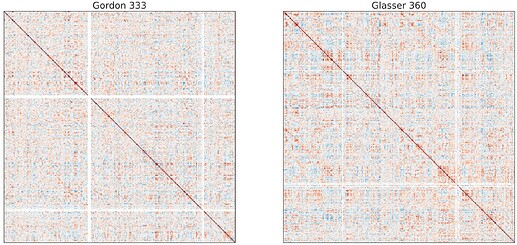
Some screen shots from one of the subjects:
And these irregular patterns (white band) occur consistently across different subjects and different task, that is, occur at similar ROIs.
I want to know whether it is related to wrong registration or something else? And any suggestions to fix this problem? I am not so familiar with neuroimages, but I will try my best to provide any other information if needed.
Thank you!