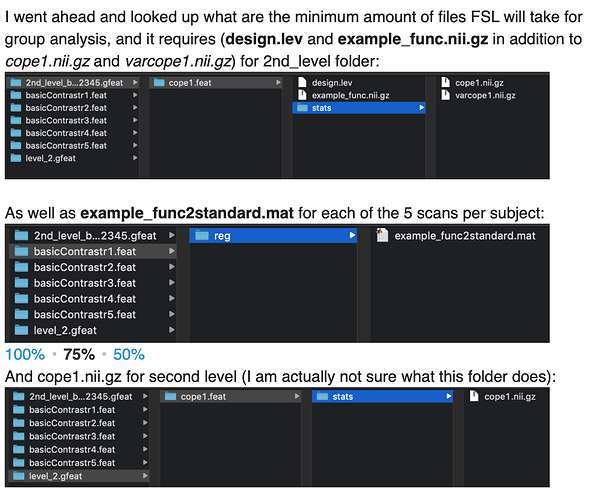
Hi,
I inherited a nipype pipeline that god moved to the local university server after it has been set up and was able to do within subject analysis and between subject analysis (I don’t even have imaging or programming background, don’t ask me how I run this pipeline on HPC, but I eventually did).
Now I need to do BETWEEN GROUP ANALYSIS and I don’t have the pipeline set up for it.
I have at least two options:
 1st OPTION. Use 2nd level files (copes and varcopes) in FSL and try to run it manually.
1st OPTION. Use 2nd level files (copes and varcopes) in FSL and try to run it manually.
Problem: FSL is very peaky about its FEAT directories, and is asking for some other files, that I don’t seem to have.
QUESTION: Is there a way around it? where I can just feed copes and varcopes files into between group analysis in FEAT?
 2nd OPTION: Figure out how to run it using nipype.
2nd OPTION: Figure out how to run it using nipype.
Problem: I barely know any Python and totally not sure I fully understand Nipype, and people from whom I inherited all gone. I need to be able to talk about it with someone to be able to solve this.
I would like to attach the files for 2nd and 3d level processing in hopes that it might be useful, but as a new user I can’t do this, so I am trying to put this code at the end of this message.
QUESTION: Can somebody help me with this based on the existing files (or at least point me in a direction of useful resources)?
I would be forever grateful for ANY help with this!
I also will be more than happy to clarify any questions.
Thank you!
Here is the code (with permission):
for PipelineConfig_001v_MLERank.py
# -*- coding: utf-8 -*-
"""
Created on Tue Aug 26 14:06:38 2014
@author: Dalton
"""
"""
=========
Imports
=========
"""
import os # system functions
import nipype.interfaces.fsl as fsl # fsl
"""
==============
Configurations
==============
"""
# for calcualting core-count
CPU_Count = 20
data_dir = "/home/rcf-proj/ib2/fmri/New_Batch"
fsl_dir = "/home/rcf-proj/ib2/fmri/fsl"
#set output file format to compressed NIFTI.
fsl.FSLCommand.set_default_output_type('NIFTI_GZ')
preproc_folder = "PreProcessed/"
models_folder = "Models/"
event_file_folder = "EventFiles/"
within_subj_results_folder = "FFX_Results/"
between_subj_results_folder = "MFX_Results/"
working_folder = "WorkingDir/"
crash_report_folder = "crashdumps/"
models = [
# "Model_001_MLERank"
# "Model_001_Rank",
# "Model_001_Rank_Offset",
# "Model_001_Rank_Offset_Split",
# "Model_001_S",
# "Model_001_S_RT",
"Model_001v_MLERank",
# "Model_001v_Rank",
# "Model_001v_S",
# "Model_001v_S_Chosen",
#etc.
]
# Templates
mfxTemplateBrain = fsl_dir + '/data/standard/MNI152_T1_2mm.nii.gz'
strippedmfxTemplateBrain= fsl_dir + '/data/standard/MNI152_T1_2mm_brain.nii.gz'
mniConfig = fsl_dir + '/etc/flirtsch/T1_2_MNI152_2mm.cnf'
mniMask = fsl_dir + '/data/standard/MNI152_T1_2mm_brain_mask_dil.nii.gz'
#List of functional scans
func_scan= [1,2,3,4,5]
# subject directories
subject_list = [
'SID3301', 'SID3303'
#etc.,
]
Here is the code for FirstLevelandWithinSubject_001v_MLERank.py
# -*- coding: utf-8 -*-
"""
Created on Mon Aug 11 12:42:49 2014
@author: Dalton
"""
#!/usr/bin/env python
# emacs: -*- mode: python; py-indent-offset: 4; indent-tabs-mode: nil -*-
# vi: set ft=python sts=4 ts=4 sw=4 et:
"""
=========
Imports
=========
"""
import os # system functions
import sys
import nipype.interfaces.io as nio # Data i/o
import nipype.interfaces.fsl as fsl # fsl
import nipype.interfaces.utility as util # utility
import nipype.pipeline.engine as pe # pypeline engine
import nipype.algorithms.modelgen as model # model generation
import nipype.algorithms.rapidart as ra # artifact detection
from nipype import LooseVersion # for simplifying versions
import json
import PipelineConfig_001v_MLERank as PC
# These two lines enable debug mode
from nipype import config
config.enable_debug_mode()
"""
==============
Configurations
==============
"""
#This should be the only thing you have to set
modelName = "Model_001v_MLERank"
from PipelineConfig_001v_MLERank import *
# Bring in the path names from the configureation file
data_dir, preProcDir, ev_dir, withinSubjectResults_dir, betweenSubjectResults_dir, workingdir,crashRecordsDir = configPaths(modelName)
sys.path.append("/home/rcf-proj/ib2/fmri/New_Batch/Models/" + modelName)
from Contrasts import *
# Get the FSL version code
version = 0
if fsl.Info.version() and \
LooseVersion(fsl.Info.version()) > LooseVersion('5.0.6'):
version = 507
"""
=========
Functions
=========
"""
#function to pick the first file from a list of files
def pickfirst(files):
if isinstance(files, list):
return files[0]
else:
return files
#function to return the 1 based index of the middle volume
def getmiddlevolume(func):
from nibabel import load
funcfile = func
if isinstance(func, list):
funcfile = func[0]
_,_,_,timepoints = load(funcfile).get_shape()
return (timepoints/2)-1
#function to get the scaling factor for intensity normalization
def getinormscale(medianvals):
return ['-mul %.10f'%(10000./val) for val in medianvals]
#function to get 10% of the intensity
def getthreshop(thresh):
return '-thr %.10f -Tmin -bin'%(0.1*thresh[0][1])
#functions to get the brightness threshold for SUSAN
def getbtthresh(medianvals):
return [0.75*val for val in medianvals]
def getusans(x):
return [[tuple([val[0],0.75*val[1]])] for val in x]
#Function to Sort Copes
def sort_copes(files):
numelements = len(files[0])
outfiles = []
for i in range(numelements):
outfiles.insert(i,[])
for j, elements in enumerate(files):
outfiles[i].append(elements[i])
return outfiles
def num_copes(files):
return len(files)
def within_subj_Dir(model_name):
withinSubjectResults_dir = '/home/rcf-proj/ib2/fmri/New_Batch/Models/' + model_name + "/FFX_Results/"
return withinSubjectResults_dir
def contrast_decode(cont_file):
import json
cont_string = open(cont_file)
cont = json.load(cont_string)
return cont
"""
======================
Configure Directoriess
======================
"""
model_name = PC.models[0]
model_folder = model_name + "/"
# Bring in the path names from the configureation file
data_dir = PC.data_dir
workingdir = PC.data_dir + PC.working_folder
crashRecordsDir = workingdir + PC.crash_report_folder
# from Contrasts import contrasts
"""
======================
model fitting workflow
======================
NODES
"""
#Master Node
modelfit = pe.Workflow(name='modelfit')
#generate design information
modelspec = pe.Node(interface=model.SpecifyModel(input_units = PC.input_units,
time_repetition = PC.TR,
high_pass_filter_cutoff = PC.hpcutoff),
name="modelspec")
#generate a run specific fsf file for analysis
level1design = pe.Node(interface=fsl.Level1Design(interscan_interval = PC.TR,
bases = {'dgamma':{'derivs': True}},
# contrasts = contrasts,
model_serial_correlations = True),
name="level1design")
#generate a run specific mat file for use by FILMGLS
modelgen = pe.MapNode(interface=fsl.FEATModel(), name='modelgen',
iterfield = ['fsf_file', 'ev_files'])
if version < 507:
#estomate Model
modelestimate = pe.MapNode(interface=fsl.FILMGLS(smooth_autocorr=True,
mask_size=5,
threshold=1000),
name='modelestimate',
iterfield = ['design_file',
'in_file'])
#estimate contrasts
conestimate = pe.MapNode(interface=fsl.ContrastMgr(), name='conestimate',
iterfield = ['tcon_file','param_estimates',
'sigmasquareds', 'corrections',
'dof_file'])
else:
#estomate Model and Contrasts
modelestimate = pe.MapNode(interface=fsl.FILMGLS(smooth_autocorr=True,
mask_size=5,
threshold=1000),
name='modelestimate',
iterfield = ['design_file',
'in_file',
'tcon_file'])
'''
CONNECTIONS
'''
if version < 507:
modelfit.connect([
(modelspec,level1design,[('session_info','session_info')]),
(level1design,modelgen,[('fsf_files', 'fsf_file'),
('ev_files', 'ev_files')]),
(modelgen,modelestimate,[('design_file','design_file')]),
(modelgen,conestimate,[('con_file','tcon_file')]),
(modelestimate,conestimate,[('param_estimates','param_estimates'),
('sigmasquareds', 'sigmasquareds'),
('corrections','corrections'),
('dof_file','dof_file')]),
])
else:
modelfit.connect([
(modelspec,level1design,[('session_info','session_info')]),
(level1design,modelgen,[('fsf_files', 'fsf_file'),
('ev_files', 'ev_files')]),
(modelgen,modelestimate,[('design_file','design_file'),
('con_file','tcon_file')]),
])
"""
======================
fixed-effects workflow
======================
NODES
"""
# Master Node
fixed_fx = pe.Workflow(name='fixedfx')
#merge the copes and varcopes for each condition
copemerge = pe.MapNode(interface=fsl.Merge(dimension='t'),
iterfield=['in_files'],
name="copemerge")
varcopemerge = pe.MapNode(interface=fsl.Merge(dimension='t'),
iterfield=['in_files'],
name="varcopemerge")
#level 2 model design files (there's one for each condition of each subject)
level2model = pe.Node(interface=fsl.L2Model(),
name='l2model')
#estimate a second level model
flameo = pe.MapNode(interface=fsl.FLAMEO(run_mode='fe',
mask_file = PC.mniMask), name="flameo",
iterfield=['cope_file','var_cope_file'])
# ROI maskes
ROIs = pe.MapNode(interface = fsl.ApplyMask(),
name ='ROIs',
iterfield=['in_file'],
iterables = ('mask_file',PC.ROI_Masks))
'''
Connections
'''
fixed_fx.connect([(copemerge,flameo,[('merged_file','cope_file')]),
(varcopemerge,flameo,[('merged_file','var_cope_file')]),
(level2model,flameo, [('design_mat','design_file'),
('design_con','t_con_file'),
('design_grp','cov_split_file')]),
(flameo, ROIs, [('tstats', 'in_file')]),
])
"""
=======================
Within-Subject workflow
=======================
NODES
"""
#Master NODE
withinSubject = pe.Workflow(name='withinSubject')
"""
CONNECTIONS
"""
if version < 507:
withinSubject.connect([(modelfit, fixed_fx,[(('conestimate.copes', sort_copes),'copemerge.in_files'),
(('conestimate.varcopes', sort_copes),'varcopemerge.in_files'),
(('conestimate.copes', num_copes),'l2model.num_copes'),
])
])
else:
withinSubject.connect([(modelfit, fixed_fx,[(('modelestimate.copes', sort_copes),'copemerge.in_files'),
(('modelestimate.varcopes', sort_copes),'varcopemerge.in_files'),
(('modelestimate.copes', num_copes),'l2model.num_copes'),
])
])
"""
=============
META workflow
=============
NODES
"""
# Master NODE
masterpipeline = pe.Workflow(name= "MasterWorkfow")
masterpipeline.base_dir = workingdir + '/FFX'
masterpipeline.config = {"execution": {"crashdump_dir":crashRecordsDir}}
# Set up inforsource to iterate over 'subject_id's
modle_subj_source = pe.Node(interface=util.IdentityInterface(fields=['model_name', 'subject_id']),
name="modle_subj_source")
modle_subj_source.iterables = [('subject_id', PC.subject_list),
('model_name', PC.models)]
# The datagrabber finds all of the files that need to be run and makes sure that
# they get to the right nodes at the begining of the protocol.
datasource = pe.Node(interface=nio.DataGrabber(infields=['model_name', 'subject_id'],
outfields=['func', 'art', 'evs', 'cont']),
name = 'datasource')
datasource.inputs.base_directory = data_dir
datasource.inputs.template = '*'
datasource.inputs.field_template= dict(func= '/home/rcf-proj/ib2/fmri/New_Batch/Sink_PreProc/%s/func2MNI/out_file/*.nii*',
art = '/home/rcf-proj/ib2/fmri/New_Batch/Sink_PreProc/%s/art/outlier_files/*.txt',
evs = '/home/rcf-proj/ib2/fmri/New_Batch/Models/%s/EventFiles/%s/RUN%d/*.txt',
cont= '/home/rcf-proj/ib2/fmri/New_Batch/Models/%s/EventFiles/contrasts.json'
)
datasource.inputs.template_args = dict(func= [['subject_id']],
art = [['subject_id']],
evs = [['model_name', 'subject_id', PC.func_scan]],
cont= [['model_name']])
datasource.inputs.sort_filelist = True
#DataSink --- stores important outputs
datasink = pe.Node(interface=nio.DataSink(parameterization = True), # This line keeps the DataSink from adding an aditional level to the directory, I have no Idea why this works.
name="datasink")
datasink.inputs.substitutions = [('_subject_id_', ''),
('_flameo', 'contrast')]
"""
CONNECTIONS
"""
masterpipeline.connect([(modle_subj_source, datasource, [('subject_id', 'subject_id'),
('model_name', 'model_name')]),
(datasource, withinSubject, [('evs', 'modelfit.modelspec.event_files'),
('func', 'modelfit.modelspec.functional_runs'),
('func', 'modelfit.modelestimate.in_file'),
('art', 'modelfit.modelspec.outlier_files'),
(('cont', contrast_decode), 'modelfit.level1design.contrasts')
]),
(modle_subj_source, datasink, [('subject_id', 'container'),
(('model_name', within_subj_Dir),'base_directory')]),
# (modle_subj_source, masterpipeline, [(('model_name', working_Dir),'base_dir'),
# (('model_name', crash_Dir),'config')])
])
#DataSink Connections -- These are put with the meta flow becuase the dataSink
# reaches in to a lot of deep places, but it is not of
# those places; hence META.
withinSubject.connect([(modelfit,datasink,[('modelestimate.param_estimates','regressorEstimates')]),
(modelfit,datasink,[('level1design.fsf_files', 'fsf_file')]),
(fixed_fx,datasink,[('flameo.tstats','tstats'),
('flameo.copes','copes'),
('flameo.var_copes','varcopes'),
]),
(ROIs, datasink,[('out_file','ROIs')])
])
"""
====================
Execute the pipeline
====================
"""
#outgraph = masterpipeline.run(plugin='MultiProc', plugin_args={'n_procs':1, 'memory_gb':30})
#outgraph = masterpipeline.run(plugin='MultiProc', plugin_args={'n_procs': 14})
outgraph = masterpipeline.run(plugin='Linear', plugin_args={'n_procs': 1})
Here is adapted code for BetweenSubject_001v_MLERank.py
"""
Created on Tue Aug 12 10:38:27 2014
@author: Dalton
"""
#!/usr/bin/env python
# emacs: -*- mode: python; py-indent-offset: 4; indent-tabs-mode: nil -*-
# vi: set ft=python sts=4 ts=4 sw=4 et:
"""
=========
Imports
=========
"""
import os # system functions
import sys
import nipype.interfaces.io as nio # Data i/o
import nipype.interfaces.fsl as fsl # fsl
import nipype.pipeline.engine as pe # pypeline engine
# from nipype import config
# config.enable_debug_mode()
"""
==============
Configurations
==============
"""
#This should be the only thing you have to set
modelName = "Model_001v_MLERank"
from PiplineConfig_BtwSubj_001v_MLERank import *
# Bring in the path names from the configureation file
data_dir, preProcDir, ev_dir, withinSubjectResults_dir, betweenSubjectResults_dir, workingdir,crashRecordsDir = configPaths(modelName)
# This needs to point to the code directory Let's see if the ~ works in this context
sys.path.append("../Models/" + modelName)
from Contrasts import *
"""
=========
Functions
=========
"""
#Function to Sort Copes
def sort_copes(files):
numelements = len(files[0])
outfiles = []
for i in range(numelements):
outfiles.insert(i,[])
for j, elements in enumerate(files):
outfiles[i].append(elements[i])
return outfiles
def num_copes(files):
return len(files)
def within_subj_Dir(model_name):
withinSubjectResults_dir = '/data/Models/' + model_name + "/FFX_Results/"
return withinSubjectResults_dir
def between_subj_Dir(model_name):
betweenSubjectResults_dir = '/data/Models/' + model_name + "/MFX_Results/"
return betweenSubjectResults_dir
def contrast_decode(cont_file):
import json
cont_string = open(cont_file)
cont = json.load(cont_string)
return cont
def contrast_counter(cont_file):
import json
cont_string = open(cont_file)
cont = json.load(cont_string)
return cont
'''
==============
META NODES
==============
'''
# MASTER node
masterpipeline = pe.Workflow(name= "MasterWorkfow")
masterpipeline.base_dir = workingdir + '/MFX'
masterpipeline.config = {"execution": {"crashdump_dir":crashRecordsDir}}
# 2nd level dataGrabber
contrast_ids = range(0,len(contrasts))
l2source = pe.Node(nio.DataGrabber(infields=['con'],
outfields=['copes','varcopes']),
name="l2source")
l2source.inputs.base_directory = withinSubjectResults_dir
l2source.inputs.template = '*'
l2source.inputs.field_template= dict(copes= '%s/copes/*%s/contrast%d/cope1.nii.gz',
varcopes= '%s/varcopes/*%s/contrast%d/varcope1.nii.gz')
l2source.inputs.template_args = dict(copes= [[subject_list,subject_list,'con']],
varcopes= [[subject_list,subject_list,'con']]
)
# iterate over all contrast images
l2source.iterables = [('con',contrast_ids)]
l2source.inputs.sort_filelist = True
# DataSink
#DataSink --- stores important outputs
MFXdatasink = pe.Node(interface=nio.DataSink(base_directory= betweenSubjectResults_dir,
parameterization = True # This line keeps the DataSink from adding an aditional level to the directory, I have no Idea why this works.
),
name="datasink")
MFXdatasink.inputs.substitutions = [('_subject_id_', ''),
('_flameo', 'contrast')]
'''
==================
Second Level Nodes
==================
'''
#merge the copes and varcopes for each condition
copemerge = pe.Node(interface=fsl.Merge(dimension='t'),
name="copemerge")
varcopemerge = pe.Node(interface=fsl.Merge(dimension='t'),
name="varcopemerge")
#level 2 model design files (there's one for each contrast)
level2model = pe.Node(interface=fsl.L2Model(),
name='l2model')
#estimate a Random-FX level model
flameo = pe.MapNode(interface=fsl.FLAMEO(run_mode='flame1',
mask_file = mniMask),
name="flameo",
iterfield=['cope_file','var_cope_file'])
## z=1.645 was changed to z=3.1
thresholdPositive = pe.MapNode(interface=fsl.Threshold(thresh = 3.1,
direction = 'below'),
name = 'thresholdPositive',
iterfield=['in_file'])
## z=1.645 was changed to z=3.1
thresholdNegative = pe.MapNode(interface=fsl.Threshold(thresh = 3.1,
direction = 'above'),
name = 'thresholdNegative',
iterfield=['in_file'])
thresholdCombined = pe.MapNode(interface=fsl.BinaryMaths(operation = 'add'),
name = 'thresholdCombined',
iterfield=['in_file', 'operand_file'])
ROIs = pe.MapNode(interface = fsl.ApplyMask(),
name ='ROIs',
iterfield=['in_file'],
iterables = ('mask_file',ROI_Masks))
'''
===========
Connections
===========
'''
masterpipeline.connect([(l2source,copemerge,[('copes','in_files')]),
(l2source,level2model,[(('copes', num_copes),'num_copes')]),
(l2source,varcopemerge,[('varcopes','in_files')])
])
masterpipeline.connect([(copemerge,flameo,[('merged_file','cope_file')]),
(varcopemerge,flameo,[('merged_file','var_cope_file')]),
(level2model,flameo, [('design_mat','design_file'),
('design_con','t_con_file'),
('design_grp','cov_split_file')]),
])
masterpipeline.connect([(flameo,thresholdPositive,[('zstats','in_file')]),
(flameo,thresholdNegative,[('zstats','in_file')]),
(thresholdPositive,thresholdCombined,[('out_file','in_file')]),
(thresholdNegative,thresholdCombined,[('out_file','operand_file')]),
(flameo,ROIs,[('zstats','in_file')])])
masterpipeline.connect([(flameo,MFXdatasink,[('copes','copes'),
('tstats','tstats'),
('var_copes','var_copes'),
('zstats','zstats'),
]),
(thresholdPositive,MFXdatasink,[('out_file','thresholdedPositive')]),
(thresholdNegative,MFXdatasink,[('out_file','thresholdedNegative')]),
(thresholdCombined,MFXdatasink,[('out_file','thresholdedCombined')]),
(ROIs,MFXdatasink,[('out_file','ROIs')])
])
if __name__ == '__main__':
# masterpipeline.write_graph(graph2use='hierarchical')
# masterpipeline.run()
outgraph = masterpipeline.run(plugin='MultiProc', plugin_args={'n_procs':1, 'memory_gb':30})
Here is adapted code for PiplineConfig_BtwSubj_001v_MLERank.py:
# -*- coding: utf-8 -*-
"""
Created on Tue Aug 26 14:06:38 2014
@author: Dalton
"""
"""
=========
Imports
=========
"""
import os # system functions
import nipype.interfaces.fsl as fsl # fsl
"""
==============
Configurations
==============
"""
# for calcualting core-count
CPU_Count = 20
data_dir = "/home/rcf-proj/ib2/fmri/New_Batch"
fsl_dir = "/home/rcf-proj/ib2/fmri/fsl"
#set output file format to compressed NIFTI.
fsl.FSLCommand.set_default_output_type('NIFTI_GZ')
preproc_folder = "PreProcessed/"
models_folder = "Models/"
event_file_folder = "EventFiles/"
within_subj_results_folder = "FFX_Results/"
between_subj_results_folder = "MFX_Results/"
working_folder = "WorkingDir/"
crash_report_folder = "crashdumps/"
models = [
# "Model_001_MLERank"
# "Model_001_Rank",
# "Model_001_Rank_Offset",
# "Model_001_Rank_Offset_Split",
# "Model_001_S",
# "Model_001_S_RT",
"Model_001v_MLERank",
# "Model_001v_Rank",
# "Model_001v_S",
# "Model_001v_S_Chosen",
#etc.
]
# Templates
mfxTemplateBrain = fsl_dir + '/data/standard/MNI152_T1_2mm.nii.gz'
strippedmfxTemplateBrain= fsl_dir + '/data/standard/MNI152_T1_2mm_brain.nii.gz'
mniConfig = fsl_dir + '/etc/flirtsch/T1_2_MNI152_2mm.cnf'
mniMask = fsl_dir + '/data/standard/MNI152_T1_2mm_brain_mask_dil.nii.gz'
#List of functional scans
func_scan= [1,2,3,4,5]
# subject directories
subject_list = [
'SID3301', 'SID3303', #etc.
]
# Paths
def configPaths(modelName = "PreProcessing",data_dir = data_dir):
# Wthere the input data comes from
preProcDir = data_dir + "/PreProcessed"
ev_dir = data_dir + "/Models/" + modelName + "/EventFiles"
# Where the outputs goes
withinSubjectResults_dir = data_dir + "/Models/" + modelName + "/FFX_Results"
betweenSubjectResults_dir= data_dir + "/Models/" + modelName + "/MFX_Results"
# Working Directory
workingdir = data_dir + "/WorkingDir/" + modelName
# Crash Records
crashRecordsDir = workingdir + "/crashdumps"
return data_dir, preProcDir, ev_dir, withinSubjectResults_dir, betweenSubjectResults_dir, workingdir,crashRecordsDir
#ModelSettings
input_units = 'secs'
hpcutoff = 120.
TR = 2.
# ROI Masks
ROI_Masks = [
# data_dir + '/ROIs/HOMiddleFrontalGyrus.nii.gz',
# data_dir + '/ROIs/lAG.nii.gz',
'/home/rcf-proj/ib2/fmri/ROIs/lIPS.nii.gz',
'/home/rcf-proj/ib2/fmri/ROIs/rIPS.nii.gz',
'/home/rcf-proj/ib2/fmri/ROIs/rLingual.nii.gz',
'/home/rcf-proj/ib2/fmri/ROIs/ACC.nii.gz',
]