Hi, I think I found an error in the online version of the BIDS validator. Below is the error report I got. As you can see, there is an error because of the presence of the file “/event.json”. At the same time there are warnings due to “*_events.tsv” files without dictionary files associated to them. As a remedy, it is propose to add an JSON file with différent choice of naming… including “/events.json”!
Finally, when I created two files: “/task-localizer.json” and “/task-localizerMars.json” (corresponding to the two tasks present for this project), all the errors and warnings disappeared.
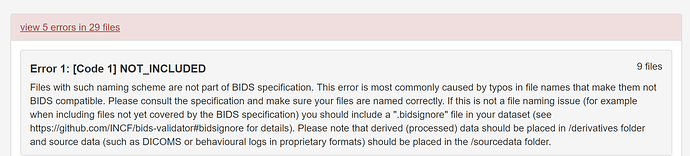
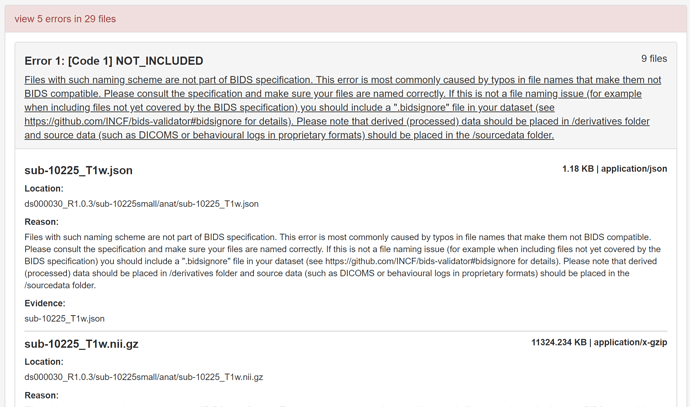
File Path: Files with such naming scheme are not part of BIDS specification. This error is most commonly caused by typos in file names that make them not BIDS compatible. Please consult the specification and make sure your files are named correctly. If this is not a file naming issue (for example when including files not yet covered by the BIDS specification) you should include a “.bidsignore” file in your dataset (see https://github.com/bids-standard/bids-validator#bidsignore for details). Please note that derived (processed) data should be placed in /derivatives folder and source data (such as DICOMS or behavioural logs in proprietary formats) should be placed in the /sourcedata folder.
Type: Error
File: events.json
Location: RemiLocaMars/events.json
Reason: Files with such naming scheme are not part of BIDS specification. This error is most commonly caused by typos in file names that make them not BIDS compatible. Please consult the specification and make sure your files are named correctly. If this is not a file naming issue (for example when including files not yet covered by the BIDS specification) you should include a ".bidsignore" file in your dataset (see https://github.com/bids-standard/bids-validator#bidsignore for details). Please note that derived (processed) data should be placed in /derivatives folder and source data (such as DICOMS or behavioural logs in proprietary formats) should be placed in the /sourcedata folder.
Evidence: events.json
======================================================
File Path: Tabular file contains custom columns not described in a data dictionary
Type: Warning
File: sub-01_task-localizerMars_acq-mb_events.tsv
Location: RemiLocaMars/sub-01/func/sub-01_task-localizerMars_acq-mb_events.tsv
Reason: Tabular file contains custom columns not described in a data dictionary
Evidence: Columns: expected_response, given_response not defined, please define in: /task-localizerMars_acq-mb_events.json, /task-localizerMars_events.json,/events.json,/sub-01/sub-01_task-localizerMars_acq-mb_events.json,/sub-01/sub-01_task-localizerMars_events.json,/sub-01/sub-01_events.json,/sub-01/func/sub-01_task-localizerMars_acq-mb_events.json,/sub-01/func/sub-01_task-localizerMars_events.json,/sub-01/func/sub-01_events.json
Type: Warning
File: sub-01_task-localizer_acq-mb_events.tsv
Location: RemiLocaMars/sub-01/func/sub-01_task-localizer_acq-mb_events.tsv
Reason: Tabular file contains custom columns not described in a data dictionary
Evidence: Columns: expected_response, given_response not defined, please define in: /task-localizer_acq-mb_events.json, /task-localizer_events.json,/events.json,/sub-01/sub-01_task-localizer_acq-mb_events.json,/sub-01/sub-01_task-localizer_events.json,/sub-01/sub-01_events.json,/sub-01/func/sub-01_task-localizer_acq-mb_events.json,/sub-01/func/sub-01_task-localizer_events.json,/sub-01/func/sub-01_events.json
Type: Warning
File: sub-01_task-localizer_acq-sb_events.tsv
Location: RemiLocaMars/sub-01/func/sub-01_task-localizer_acq-sb_events.tsv
Reason: Tabular file contains custom columns not described in a data dictionary
Evidence: Columns: expected_response, given_response not defined, please define in: /task-localizer_acq-sb_events.json, /task-localizer_events.json,/events.json,/sub-01/sub-01_task-localizer_acq-sb_events.json,/sub-01/sub-01_task-localizer_events.json,/sub-01/sub-01_events.json,/sub-01/func/sub-01_task-localizer_acq-sb_events.json,/sub-01/func/sub-01_task-localizer_events.json,/sub-01/func/sub-01_events.json
Type: Warning
File: sub-01_task-localizer_echo-1_events.tsv
Location: RemiLocaMars/sub-01/func/sub-01_task-localizer_echo-1_events.tsv
Reason: Tabular file contains custom columns not described in a data dictionary
Evidence: Columns: expected_response, given_response not defined, please define in: /task-localizer_echo-1_events.json, /task-localizer_events.json,/events.json,/sub-01/sub-01_task-localizer_echo-1_events.json,/sub-01/sub-01_task-localizer_events.json,/sub-01/sub-01_events.json,/sub-01/func/sub-01_task-localizer_echo-1_events.json,/sub-01/func/sub-01_task-localizer_events.json,/sub-01/func/sub-01_events.json
Type: Warning
File: sub-01_task-localizer_echo-2_events.tsv
Location: RemiLocaMars/sub-01/func/sub-01_task-localizer_echo-2_events.tsv
Reason: Tabular file contains custom columns not described in a data dictionary
Evidence: Columns: expected_response, given_response not defined, please define in: /task-localizer_echo-2_events.json, /task-localizer_events.json,/events.json,/sub-01/sub-01_task-localizer_echo-2_events.json,/sub-01/sub-01_task-localizer_events.json,/sub-01/sub-01_events.json,/sub-01/func/sub-01_task-localizer_echo-2_events.json,/sub-01/func/sub-01_task-localizer_events.json,/sub-01/func/sub-01_events.json
Type: Warning
File: sub-01_task-localizer_echo-3_events.tsv
Location: RemiLocaMars/sub-01/func/sub-01_task-localizer_echo-3_events.tsv
Reason: Tabular file contains custom columns not described in a data dictionary
Evidence: Columns: expected_response, given_response not defined, please define in: /task-localizer_echo-3_events.json, /task-localizer_events.json,/events.json,/sub-01/sub-01_task-localizer_echo-3_events.json,/sub-01/sub-01_task-localizer_events.json,/sub-01/sub-01_events.json,/sub-01/func/sub-01_task-localizer_echo-3_events.json,/sub-01/func/sub-01_task-localizer_events.json,/sub-01/func/sub-01_events.json
======================================================