I will add a check for valid DICOM files (which is more expensive than checking file extensions)
Thank you, really appreciate it - will you let me know?
I deleted the 5 report files (with only 4 dicom tags that were throwing errors) and then I repeatedly removed the .DS_Store files from tmp_data. I then ran dicomsort tmp_data -p ‘.*’ which gave the following tree (snippet):
tmp_data
├── 001-DTI_2x2x2.2_Multishell_20ch_2020_fftscale_SBRef
│ └── EE5829F5
├── 001-Ortho Spine
│ └── EEFDA714
├── 001-Topogram 1.0 Tr20
│ └── EE13C29D
├── 002-DTI_2x2x2.2_Multishell_20ch_2020_fftscale
│ ├── EE012DD9
│ ├── EE04C8A6
│ ├── EE06DB37
│ ├── EE0B7932
…
│ ├── EEE4396D
│ ├── EEEAEC22
│ ├── EEEFFB9F
│ ├── EEF9C68A
│ └── EEFBA9A4
├── 401-Enhancement curve
│ └── EE545478
├── 501-Dose Report
│ ├── FFE86D52
│ └── FFF4FC96
├── 502-Patient Protocol
│ ├── EEE5CAF1
│ └── EEE83FBE
└── DICOM
└── 0000AC37
├── AA140CCD
│ └── AABD7737
Mhhh, you are merging all the sessions in this way, not what you want. So it’s back to DICOMDIR. The biggest problem is that all your anonymized subjects have the same name, i.e. “Anonymous”. I guess I could make an option to use something different for the subject folder than PatientName when dealing with DICOMDIR (presumably PatientID)
Can you check if the PatientID field is not anonymized?
Yes - the Patient ID field is not anonymised, e.g. ANON(with 9 nine letters/numbers afterwards).
The DICOMDIR support in bidscoin is limited because I don’t have any such data to test things, except for the multi-patient DICOMDIR test-data from pydicom. What I would need is single-patient DICOMDIR data (such as you have) to develop a more robust sorting. I’ll see if I can find any
Question, are there dicom files in tmp_data\Anonymous\01-? I suppose this is the first of your 6 sessions? Can you tell me what 001-Topogram 1.0 Tr20 etc is? To what session does it belong?
Hi Marcel,
To recap, I’ve run:
- cp -r patient-01 tmp_data
- then manually removed the 5 report files without SeriesDescription that threw errors (by the way - I have looked at these again, and although they do not have SeriesDescription, they do have more than just 4 dicomtags (I think I had a search filter for series) contrary to what I said yesterday).
- dicomsort tmp_data
This gives warnings for the deleted files - WARNING | The referenced SOP Instance for the directory record at offset 864 does not exist …
The tree now looks like this (discontinuous snippets as too long to include the tree):
tmp_data
├── Anonymous
│ ├── 01-
│ │ ├── 002-t2_tse_tra_448
│ │ │ ├── EE02A56B
│ │ │ ├── EE10912D
│ │ │ ├── EE1451F7
│ │ │ ├── EE16A62C
│ │ │ ├── EE1C4B1C
│ │ │ ├── EE1DAD46
│ │ │ ├── EE20FAD0
│ │ │ ├── EE2B176A
…
│ │ ├── EEFCC461
│ │ └── EEFF5F03
│ ├── 02-
│ │ ├── 004-pCASL_BL1800_PLD1500_con
│ │ │ ├── EE276E3C
│ │ │ ├── EE2B9118
│ │ │ ├── EE2BD130
│ │ │ ├── EE37474C
│ │ │ ├── EE3A099A
│ │ │ ├── EE3BC6D5
│ │ │ ├── EE4128D7
…
│ │ └── EEFBA9A4
│ ├── 03-
│ │ ├── 001-DTI_2x2x2.2_Multishell_20ch_2020_fftscale_SBRef
│ │ │ └── EE5829F5
│ │ ├── 002-DTI_2x2x2.2_Multishell_20ch_2020_fftscale
│ │ │ ├── EE012DD9
│ │ │ ├── EE04C8A6
It has created folders for all 6 sessions and they have DICOMS in them.
I think it didn’t give this tree yesterday because I hadn’t deleted the report files (without SeriesDescription), however, looking at my previous attempts to use dicomsort on Monday, I used dicomsort -f {SeriesNumber: 03 d}-{SeriesDescription} --force patient-01, and at that time, it gave a similar tree output as above (except with additional folders for the 5 radiology reports). I tried to use the -i -j in various ways as well but could not figure it out and it didn’t give any output using these.
In answer to your question, yes there are now dicom files in tmp_data\Anonymous\01 and this is the first of the 6 sessions.
I’m not sure if this updated tree helps in any way with the bidsmapping? I’m not sure how to derive the sub / ses from the current structure.
Okay, I think I’ve covered it all in my latest code now, except for your topogram folder. What is it doing there, and which session does it belong to (or is it a subfolder of 06)?
Wow! - Thank you! I have two topogram folders:
│ ├── 04-
│ │ ├── 002-Topogram 1.0 Tr20
│ │ │ ├── EE6130EB
│ │ │ └── EE6B049D
and I’ve attached the second as an image because when pasted as text here it looks like its in the wrong place
The topograms below to sessions 4 and 6, respectively.
You could perhaps try it out (I don’t have the data to test it myself) by installing the latest code from GitHub? The code should now be resilient against your DS_Store files, so you should run it on your original data (i.e. without deleting anything)
python -m venv bidscoin_dev
source bidscoin_dev/bin/activate
pip install "git+https://github.com/Donders-Institute/bidscoin"
bidsmapper sourcedata BIDS_new -s -n 'patient-' -m '*'
bidscoiner sourcedata BIDS_new
Hi Marcel,
Thank you for your help so far - really appreciate it. In summary, it works but only if I manually delete the radiology report files.
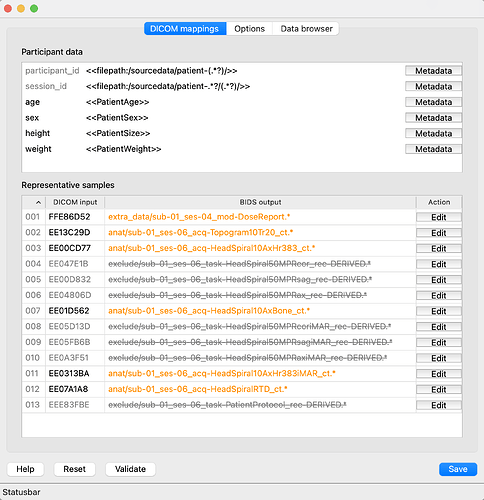
Interestingly, at first, it picks up only ses-4 and ses-6 (see screenshot). It appears that the 5 radiology report files (which are missing ‘SeriesDescription’) are blocking it from creating subfolders:
2025-03-19 18:12:06 - ERROR | Cannot create subfolders, aborting dicomsort()…
2025-03-19 18:12:06 - ERROR | Missing ‘SeriesDescription’ DICOM field specified in the ‘{SeriesNumber:03d}-{SeriesDescription}’ folder/naming scheme, cannot find a safe name for: /private/var/folders/ … sourcedata/patient-01/DICOM/0000AC37/AA740734/AAB9BE77/00007A3B/FFA7A389.
Does the above suggest that it’s not the DS_Store files which are causing the problem - instead the report files?
It also gives a BIDScoiner version difference, but I don’t think this is a problem: /Users/user/.bidscoin/4.5.2.dev0/templates/bidsmap_dccn.yaml was created with version 4.5.2.dev, but this is version 4.5.2.dev0. This is normally OK but check the Changelog — BIDScoin 4.5.2.dev documentation
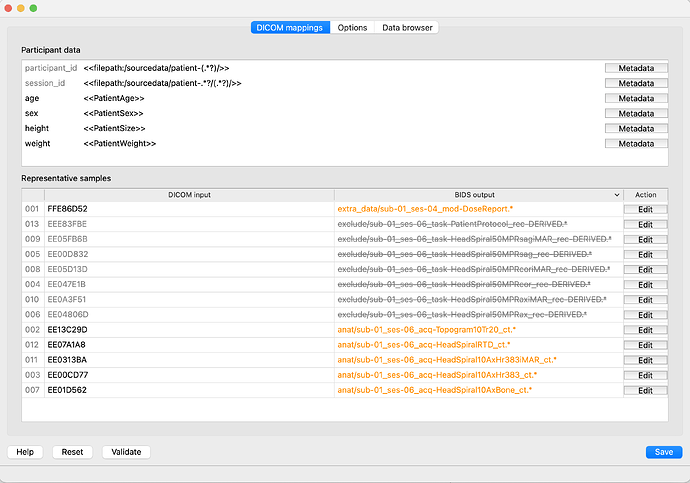
The bidseditor GUI does not seem to have full functionality, e.g. sliding the column to expand the width under ‘Representative Sample’ as before. Is this intentional because it is a developer version or an error at my end?
So, I manually deleted the files which gave the error above, and re-ran the code. It was then able to work, including bidscoiner, which gave the following output (below).
I think the BIDS output structure looks correct, however, I had to manually delete the files in order for this code to work: bidsmapper sourcedata BIDS_new -s -n ‘patient-’ -m ‘*’
I have checked the radiology reports and they all have the dicom tag (0008,0104) CodeMeaning with value ‘Radiology Report’. Do you think the block comes from the radiology reports and is there a way to exclude them based on this in order to automate the process?
sub-01
├── ses-01
│ ├── extra_data
│ │ ├── sub-01_ses-01_mod-MagImages_echo-1_GR.json
│ │ ├── sub-01_ses-01_mod-MagImages_echo-1_GR.nii.gz
│ │ ├── sub-01_ses-01_mod-PhaImages_echo-1_part-phase_GR.json
│ │ ├── sub-01_ses-01_mod-PhaImages_echo-1_part-phase_GR.nii.gz
│ │ ├── sub-01_ses-01_mod-SWIImages_echo-1_GR.json
│ │ ├── sub-01_ses-01_mod-SWIImages_echo-1_GR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV1_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV1_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV2_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mp2ragesagp3isoINV2_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopost_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopost_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopre_echo-1_GRIR.json
│ │ ├── sub-01_ses-01_mod-t1mpragesagp2isopre_echo-1_GRIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t2spcflairsag1mmiso_echo-1_SEIR.json
│ │ ├── sub-01_ses-01_mod-t2spcflairsag1mmiso_echo-1_SEIR.nii.gz
│ │ ├── sub-01_ses-01_mod-t2tsecor448_echo-1_SE.json
│ │ ├── sub-01_ses-01_mod-t2tsecor448_echo-1_SE.nii.gz
│ │ ├── sub-01_ses-01_mod-t2tsetra448_echo-1_SE.json
│ │ └── sub-01_ses-01_mod-t2tsetra448_echo-1_SE.nii.gz
│ └── sub-01_ses-01_scans.tsv
├── ses-02
│ ├── extra_data
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500con_echo-1_EP.json
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500con_echo-1_EP.nii.gz
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500diff_echo-1_EP.json
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500diff_echo-1_EP.nii.gz
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500lab_echo-1_EP.json
│ │ ├── sub-01_ses-02_mod-pCASLBL1800PLD1500lab_echo-1_EP.nii.gz
│ │ ├── sub-01_ses-02_mod-ssTE00TI1700_echo-1_EP.json
│ │ └── sub-01_ses-02_mod-ssTE00TI1700_echo-1_EP.nii.gz
│ └── sub-01_ses-02_scans.tsv
├── ses-03
│ ├── dwi
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020fftscale_sbref.json
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020fftscale_sbref.nii.gz
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020negpefftscale_sbref.json
│ │ ├── sub-01_ses-03_acq-DTI2x2x22Multishell20ch2020negpefftscale_sbref.nii.gz
│ │ ├── sub-01_ses-03_run-1_dwi.bval
│ │ ├── sub-01_ses-03_run-1_dwi.bvec
│ │ ├── sub-01_ses-03_run-1_dwi.json
│ │ ├── sub-01_ses-03_run-1_dwi.nii.gz
│ │ ├── sub-01_ses-03_run-2_dwi.bval
│ │ ├── sub-01_ses-03_run-2_dwi.bvec
│ │ ├── sub-01_ses-03_run-2_dwi.json
│ │ └── sub-01_ses-03_run-2_dwi.nii.gz
│ └── sub-01_ses-03_scans.tsv
├── ses-04
│ ├── anat
│ │ ├── sub-01_ses-04_acq-DEHeadAngio10Hr383F08_ct.json
│ │ ├── sub-01_ses-04_acq-DEHeadAngio10Hr383F08_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-DEHeadAngio10Hr383F08_ct_ROI1.nii.gz
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403A80kV_ct.json
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403A80kV_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403BSn150kV_ct.json
│ │ ├── sub-01_ses-04_acq-DEPPDEHeadAngio10Qr403BSn150kV_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-Monitoring100Br36_ct.json
│ │ ├── sub-01_ses-04_acq-Monitoring100Br36_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-Monitoring100Br36_ct_ROI1.nii.gz
│ │ ├── sub-01_ses-04_acq-PreMonitoring100Br36_ct.json
│ │ ├── sub-01_ses-04_acq-PreMonitoring100Br36_ct.nii.gz
│ │ ├── sub-01_ses-04_acq-PreMonitoring100Br36_ct_ROI1.nii.gz
│ │ ├── sub-01_ses-04_acq-Topogram10Tr20_ct.json
│ │ ├── sub-01_ses-04_acq-Topogram10Tr20_ct.nii.gz
│ │ └── sub-01_ses-04_acq-Topogram10Tr20_ct_ROI1.nii.gz
│ ├── extra_data
│ └── sub-01_ses-04_scans.tsv
├── ses-05
│ ├── extra_data
│ │ ├── sub-01_ses-05_run-10_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-10_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-11_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-11_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-12_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-12_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-1_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-1_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-2_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-2_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-3_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-3_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-4_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-4_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-5_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-5_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-6_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-6_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-7_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-7_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-8_mod-OrthoSpine.json
│ │ ├── sub-01_ses-05_run-8_mod-OrthoSpine.nii.gz
│ │ ├── sub-01_ses-05_run-9_mod-OrthoSpine.json
│ │ └── sub-01_ses-05_run-9_mod-OrthoSpine.nii.gz
│ └── sub-01_ses-05_scans.tsv
└── ses-06
├── anat
│ ├── sub-01_ses-06_acq-HeadSpiral10AxBone_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiral10AxBone_ct.nii.gz
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383_ct.nii.gz
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383iMAR_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiral10AxHr383iMAR_ct.nii.gz
│ ├── sub-01_ses-06_acq-HeadSpiralRTD_ct.json
│ ├── sub-01_ses-06_acq-HeadSpiralRTD_ct.nii.gz
│ ├── sub-01_ses-06_acq-Topogram10Tr20_ct.json
│ └── sub-01_ses-06_acq-Topogram10Tr20_ct.nii.gz
├── extra_data
└── sub-01_ses-06_scans.tsv
Thanks, that’s very useful feedback! Your 6 studies, have they been acquired in one session or did your patient really get in and out of the scanner 6 times? I’ll have a further look tomorrow to see if I can iron out the last issues…
The 6 studies happened on 3 separate dates. 4 of the studies happened on the same day, and of these, 3 studies happened at the same time (I’m basing this off the dicom metadata) which I assume means the patient the did not get out of the scanner for these 3 sessions. Is this an issue?
It’s not an issue, I was just wondering what exactly, in DICOM terms, was defining a “study” and how well this maps onto a BIDS session
Btw, I have no experience with CT, so I did not add all the CT metadata fields (from BEP024). And I suppose that dcm2niix hasn’t done that either, which means that to be BIDS compliant, you will have to fill out the metadata table yourself (sorry), either in the bidseditor, or preferably, in the template bidsmap. If you do the latter, perhaps then you can submit a pull request or send me your edits, to the benefit of everybody ![]()
Thank you, I understand. I don’t have experience with CT either and have/am mainly using MRI scans, but would use CT for lead localisation, for example.
Ok, with the latest code you should not have any issues anymore, let me know if otherwise (including GUI regressions and, especially, the radiology report errors):
source bidscoin_dev/bin/activate
pip install --force-reinstall --no-deps "git+https://github.com/Donders-Institute/bidscoin"
bidsmapper sourcedata BIDS_new -s -n 'patient-' -m '*'
bidscoiner sourcedata BIDS_new
Thanks again for the feedback, I couldn’t have fixed this without your help
Hi, I’m afraid that, as before, it only picks up ses-04 and ses-06, seemingly related to 5 files lacking ‘SeriesDescription’. Here are the errors:
INFO | >> Sorting: /var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... sourcedata/patient-01/01- (3908 files)
ERROR | Missing 'SeriesDescription' DICOM field specified in the '{SeriesNumber:03d}-{SeriesDescription}' folder/naming scheme, cannot find a safe name for: /private/var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/DICOM/0000AC37/AA5C483A/AA5BB5F3/0000A3A7/FFD51A51
ERROR | Cannot create subfolders, aborting dicomsort()...
INFO | >> Sorting: /var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/02- (217 files)
ERROR | Missing 'SeriesDescription' DICOM field specified in the '{SeriesNumber:03d}-{SeriesDescription}' folder/naming scheme, cannot find a safe name for: /private/var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/DICOM/0000AC37/AAD32921/AA448862/0000730C/FF35FD2C
ERROR | Cannot create subfolders, aborting dicomsort()...
INFO | >> Sorting: /var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/03- (729 files)
ERROR | Missing 'SeriesDescription' DICOM field specified in the '{SeriesNumber:03d}-{SeriesDescription}' folder/naming scheme, cannot find a safe name for: /private/var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/DICOM/0000AC37/AA740734/AAB9BE77/00007A3B/FFA7A389
ERROR | Cannot create subfolders, aborting dicomsort()...
INFO | >> Sorting: /var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/04- (884 files)
VERBOSE | Creating: /var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users ... /sourcedata/patient-01/04-/501-Dose Report
ERROR | Missing 'SeriesDescription' DICOM field specified in the '{SeriesNumber:03d}-{SeriesDescription}' folder/naming scheme, cannot find a safe name for: /private/var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/DICOM/0000AC37/AA140CCD/AABD7737/000098FB/FFBFB03E
ERROR | Cannot create subfolders, aborting dicomsort()...
INFO | >> Sorting: /var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/05- (15 files)
ERROR | Missing 'SeriesDescription' DICOM field specified in the '{SeriesNumber:03d}-{SeriesDescription}' folder/naming scheme, cannot find a safe name for: /private/var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/DICOM/0000AC37/AA5C6C3B/AA2EF4AD/000074A8/FFDF25C9
ERROR | Cannot create subfolders, aborting dicomsort()...
INFO | >> Sorting: /var/folders/d9/lz2pjlr50ygdpn7ddzp2fbc40000gp/T/tmpsnocgrnf/Users/ ... /sourcedata/patient-01/06- (744 files)```
The GUI, however, does seem to be working normally: