Hello,
I need to register my T1 images of animal brain to a atlas brain I found online but before that I have to match the orientation and since they are completely different MRI images of different pigs, they are in different space. How do I match the orientation and rotate it. I found the function called fslreorient2std but it Reorient an image to match the orientation of the standard template images (MNI152). If I want to just change the orientation and rotate it same as atlas how do i do it in FSL.
Hi @Sanjida ,
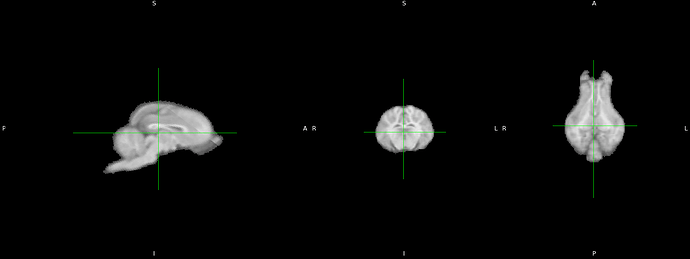
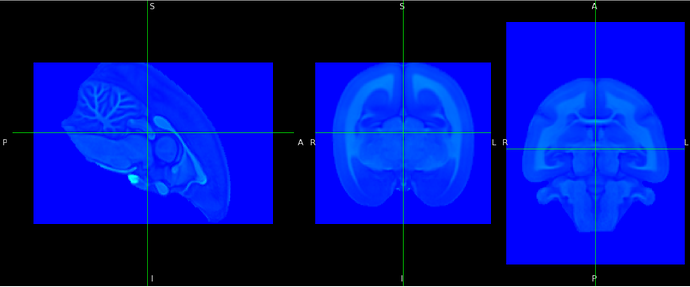
Could you show a snapshot of your T1w animal brain image in FSLeyes and one of the template space you wish to use?
The first thing is to check that the labels (L,R, P,A,I,S) are placed properly on your animal image.
It could be useful that you share also the header content of both images with fslhd.
For difficult registrations such as this, I tend to start with NiftyReg which is quite powerful.
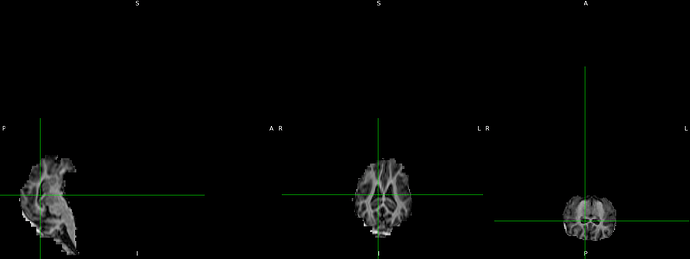
Hi @Sanjida , I see.
Your T1w image had indeed wrong labels (the anterior part of the image is pointing toward ‘S’ for instance in FSLeyes). But what is nice is that both your T1w image and the template are skull stripped which will help the registration process.
I think that if you would try reg_aladin(from NiftyReg) , you could get a decent registration in a few seconds. For other tools such as flirt and fnirt, you may need to start to manually rotate your T1w image toward the template orientation with FSLeyes “nudge” tool for example.
Hi @jsein i have done the skull stripping manually for the T1w, now i need to rotate it and match the orientation of the atlas. Can you tell me how to rotate the T1w in FSL. fslreorient2std does that but it Reorient an image to match the orientation of the standard template images (MNI152), is that a problem
Hi @Sanjida , fslreorient2std will not correct your labels R,L,A,P,S,I if there are displayed wrongly on your image.
Please look at the following thread where you should be able to find a solution that fits your needs:
Hi @jsein , I am actually doing the registration part with ANTS but before that can I just reorient image using nudge tools in FSL, should I still do the affine transformation part and load the atlas as reference image to reorient my T1w
Hi @Sanjida,
Yes, what you propose seems right: First do a nudge with FSLeyes to change the affine transformation of your image header to orient your T1w image closer to the atlas orientation. Then use ANTs to register the two images with the template being the reference of the registration.
Do you need to do a linear or non linear normalization of your T1w image in the end?
hi @jsein I will do a linear normalization since I am doing linear registration in ANTs I believe.
But could you help me please, I could rotate the image but once I apply and save it in the nudge tools option and reopen it is the same as before.
How to permanently change the orientation and save it?
What you are doing: rotate - apply and save should work. Do you save it to a new name? You can also save the affine to apply it later if you need.
You can save it in two types: FLIRT or voxel-to-world.
“FLIRT” means that you will be able to apply it with a flirt command.
“voxel-to-world” means that you will b able to apply it with fslorient -setsform command.
After “Apply” in nudge, you can verify that the affine of your image is changed by looking at the information of your image in FSLeyes ( “i” symbol on the top left ) or by typing fslhd your_image in a terminal.
When you say that it is the same as before, is it the case when you view it with FSLeyes or another viewer?
hi @jsein,
I believe i did the same. i set the angle and apply and saved in a new file. but once i open the file with FSLEYES again it is the same as before.
For the two option FLIRT and voxel- to- world which option you suggest choosing.
Also,if i try to save the affine to apply, I am asked to choose some matrix file, what am I supposed to do there.
Really appreciate your help.
Thank you,
Sanjida
Hi @Sanjida ,
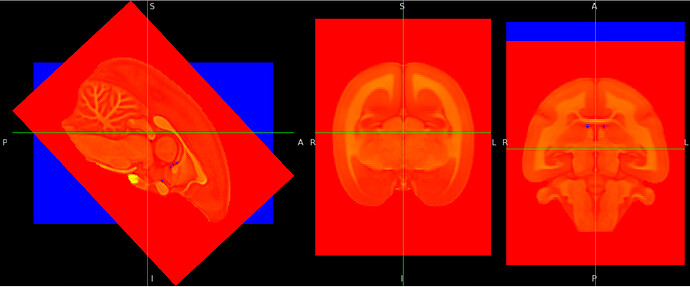
I think that you think it did not change due to the display space used by FSLeyes for your image. ![]()
Could you try to load both your original image and the image saved after “nudge”? I think they will not be aligned with each other if you applied a rotation to one of them via nudge.
You can also display your saved image in “world space” (display space: world) to see its actual orientation in the physical space.
If you look at the header of the images before and after the application of the rotation with nudge, you should see that they are indeed different.
fslhd your_image_orig.nii.gz
fslhd you_image_saved_after_nudge.nii.gz
The way to use them is different:
- The
FLIRToption saves a transformation that you can apply to your image with:
flirt -in your_original_image -ref your_original_image -out your_rotated_image -applyxfm -init your_transform_from_nudge.txt
to transform it to a rotated version of it.
- The option voxel-to-world saves the final affine of your image after nudge. To use it you have to change the affine from your original image using:
fslorient -setsform $(cat my_saved_affine_with_nudge.txt) your_image
(use a copy of your original image for this command to not loose the original orientation).
The results of both command will also be different. fslorient is just changing the header of the image and no resampling to a new grid is involved. The grid is just rotated in space. But with flirt it is different, your input image is rotated, then resampled to the reference image grid.
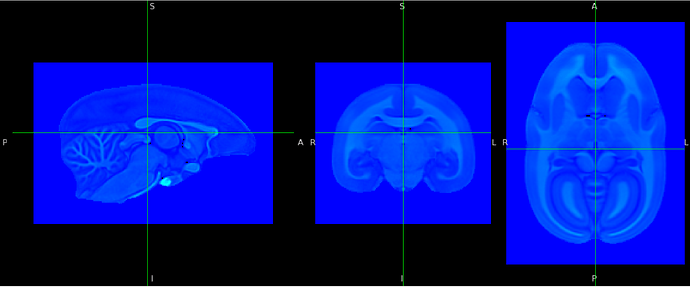
Here is an example to illustrate this: All images are displayed in “world space”.
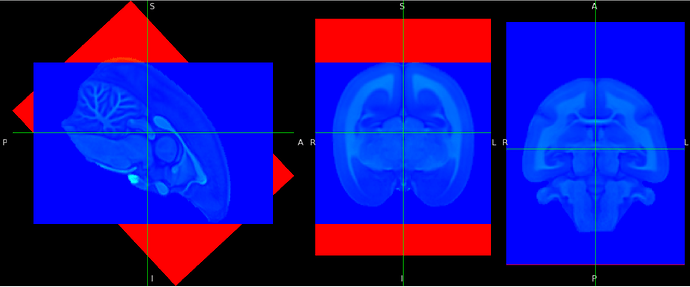
- Original image in world space. I use a colormap to show the background and see the grid of the image:
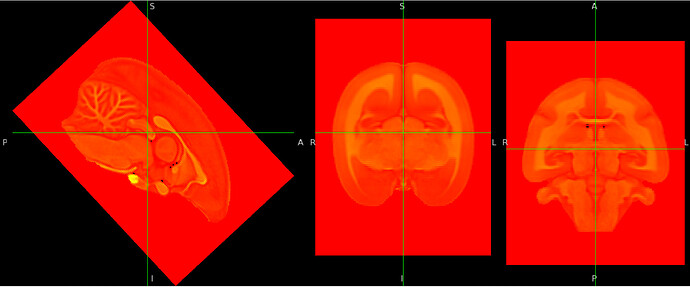
- Image to which I applied a rotation around the x axis (L-R axis) and saved it. It is the same result if I save a matrix with the option “voxel-to-world” , let’s say
nudge_affine.txtand use it this way:fslorient -setsform $(cat nudge_affine.txt) my_image.nii.gz:
- Image to which I apply with
flirtthe affine transform saved with the option “FLIRT” :
=> in both case 2. and 3., the brain have the same orientation (front tilted to the bottom) but the grid are different! Case 3 is using the original grid and the back of the brain is cut because this part of the brain moved out of the extent of the original grid!
Here two last snapshots: Case 2 on top of case 3:
And Case 3 on top of case 2:
I hope this helps understanding what you are observing!
Hi,
I am really thankful for your last help. Recently I needed help to do TBSS Analysis in Pig Brain. I am using the pig atlas from ( High-resolution magnetic resonance imaging-based atlases for the young and adolescent domesticated pig (Sus scrofa)). Here we have FA template which is needed in one step.
If I want to make it wok for our Pig scans how should I make it work since there are from different space
Thank you
Sanjida
If the template and atlas are not in the same space, you may need to coregister one space into the other with a non linear registration. There are many tools available to do so: NiftyReg, FSL, ANTS, AFNI, SPM …
I hope this helps?
Thank you. I think I can register the our FA map to atlas FA template with antstranform command i believe. What should I do exactly, I have to register all the FA map of our pig scan to the FA template?
If I understand well, what you are trying to accomplish here is the step 2 of TBSS processing, i.e. TBSS_2reg: TBSS/UserGuide - FslWiki
I never tried it, but you may try to run this step with the -t option and supply your own template as the target of the registration?