Hey,
I am running fMRIprep with ICA-AROMA and it worked perfectly for all my subjects, apart from one, for which fmriprep crashes with the following error message:
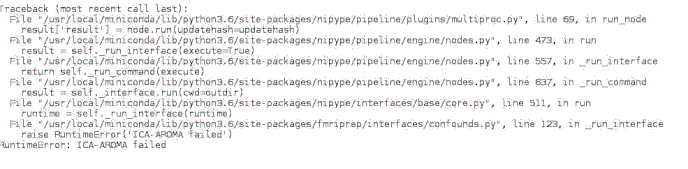
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.6/site-packages/raven/transport/threaded.py", line 165, in send_sync
super(ThreadedHTTPTransport, self).send(url, data, headers)
File "/usr/local/miniconda/lib/python3.6/site-packages/raven/transport/http.py", line 43, in send
ca_certs=self.ca_certs,
File "/usr/local/miniconda/lib/python3.6/site-packages/raven/utils/http.py", line 66, in urlopen
return opener.open(url, data, timeout)
File "/usr/local/miniconda/lib/python3.6/urllib/request.py", line 526, in open
response = self._open(req, data)
File "/usr/local/miniconda/lib/python3.6/urllib/request.py", line 544, in _open
'_open', req)
File "/usr/local/miniconda/lib/python3.6/urllib/request.py", line 504, in _call_chain
result = func(*args)
File "/usr/local/miniconda/lib/python3.6/site-packages/raven/utils/http.py", line 46, in https_open
return self.do_open(ValidHTTPSConnection, req)
File "/usr/local/miniconda/lib/python3.6/urllib/request.py", line 1320, in do_open
raise URLError(err)
urllib.error.URLError: <urlopen error [Errno -3] Temporary failure in name resolution>
['fMRIPrep running']
180930-09:17:09,261 nipype.workflow INFO:
[Node] Setting-up "fmriprep_wf.single_subject_108_wf.anat_preproc_wf.surface_recon_wf.gifti_surface_wf.get_surfaces" in "/tmp/work/fmriprep_wf/single_subject_108_wf/anat_preproc_wf/surface_recon_wf/gifti_surface_wf/get_surfaces".180930-09:17:09,261 nipype.workflow INFO:
[Node] Setting-up "fmriprep_wf.single_subject_108_wf.anat_preproc_wf.surface_recon_wf.segs_to_native_aparc_aseg.fs_datasource" in "/tmp/work/fmriprep_wf/single_subject_108_wf/anat_preproc_wf/surface_recon_wf/segs_to_native_aparc_aseg/fs_datasource".
180930-09:17:09,264 nipype.workflow INFO:
[Node] Running "fs_datasource" ("nipype.interfaces.io.FreeSurferSource")180930-09:17:09,264 nipype.workflow INFO:
[Node] Running "get_surfaces" ("nipype.interfaces.io.FreeSurferSource")
180930-09:17:09,285 nipype.workflow INFO:
[Node] Finished "fmriprep_wf.single_subject_108_wf.anat_preproc_wf.surface_recon_wf.segs_to_native_aparc_aseg.fs_datasource".
180930-09:17:09,285 nipype.workflow INFO:
[Node] Finished "fmriprep_wf.single_subject_108_wf.anat_preproc_wf.surface_recon_wf.gifti_surface_wf.get_surfaces".
180930-09:17:09,831 nipype.workflow INFO:
[Node] Setting-up "fmriprep_wf.single_subject_108_wf.anat_preproc_wf.surface_recon_wf.segs_to_native_aseg.fs_datasource" in "/tmp/work/fmriprep_wf/single_subject_108_wf/anat_preproc_wf/surface_recon_wf/segs_to_native_aseg/fs_datasource".
180930-09:17:09,835 nipype.workflow INFO:
[Node] Running "fs_datasource" ("nipype.interfaces.io.FreeSurferSource")
180930-09:17:09,855 nipype.workflow INFO:
[Node] Finished "fmriprep_wf.single_subject_108_wf.anat_preproc_wf.surface_recon_wf.segs_to_native_aseg.fs_datasource".
180930-09:17:58,82 nipype.workflow INFO:
[Node] Setting-up "fmriprep_wf.single_subject_108_wf.func_preproc_task_visualfixedlength_run_04_wf.ica_aroma_wf.ica_aroma_confound_extraction" in "/tmp/work/fmriprep_wf/single_subject_108_wf/func_preproc_task_visualfixedlength_run_04_wf/ica_aroma_wf/ica_aroma_confound_extraction".
180930-09:17:58,83 nipype.workflow INFO:
[Node] Outdated cache found for "fmriprep_wf.single_subject_108_wf.func_preproc_task_visualfixedlength_run_04_wf.ica_aroma_wf.ica_aroma_confound_extraction".
180930-09:17:58,86 nipype.workflow INFO:
[Node] Running "ica_aroma_confound_extraction" ("fmriprep.interfaces.confounds.ICAConfounds")
/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/interfaces/confounds.py:224: UserWarning: loadtxt: Empty input file: "/tmp/work/fmriprep_wf/single_subject_108_wf/func_preproc_task_visualfixedlength_run_04_wf/ica_aroma_wf/ica_aroma/out/classified_motion_ICs.txt"
motion_ic_indices = np.loadtxt(motion_ics, dtype=int, delimiter=',', ndmin=1) - 1
180930-09:17:58,97 nipype.interface WARNING:
No noise components were classified
180930-09:17:58,98 nipype.workflow WARNING:
[Node] Error on "fmriprep_wf.single_subject_108_wf.func_preproc_task_visualfixedlength_run_04_wf.ica_aroma_wf.ica_aroma_confound_extraction" (/tmp/work/fmriprep_wf/single_subject_108_wf/func_preproc_task_visualfixedlength_run_04_wf/ica_aroma_wf/ica_aroma_confound_extraction)
Traceback (most recent call last):
File "/usr/local/miniconda/bin/fmriprep", line 11, in <module>
sys.exit(main())
File "/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/cli/run.py", line 344, in main
fmriprep_wf.run(**plugin_settings)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/workflows.py", line 595, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/plugins/base.py", line 162, in run
self._clean_queue(jobid, graph, result=result))
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/plugins/base.py", line 224, in _clean_queue
raise RuntimeError("".join(result['traceback']))
RuntimeError: Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/plugins/multiproc.py", line 69, in run_node
result['result'] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py", line 471, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py", line 555, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py", line 635, in _run_command
result = self._interface.run(cwd=outdir)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/interfaces/base/core.py", line 521, in run
runtime = self._run_interface(runtime)
File "/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/interfaces/confounds.py", line 119, in _run_interface
raise RuntimeError('ICA-AROMA failed')
RuntimeError: ICA-AROMA failed
Sentry is attempting to send 1 pending error messages
Can anyone tell why it’s failing? I am on an Ubuntu system, using fmriprep v.1.1.6. This is the command I am using:
sudo docker run -it --rm -v /data/data01/BV-RT/sourcedata/:/data:ro -v /data/data01/Reece/BV-RT/fmriprep/:/out -v /data/data01/BV-RT/FS_license.txt:/opt/freesurfer/license.txt poldracklab/fmriprep:latest /data /out/out participant --participant-label 108 --fs-license-file /opt/freesurfer/license.txt --output-space template --use-aroma
Any help would be greatly appreciated,
Kristina
 I’ll submit a pull request to fmriprep, once it’s merged in, I can give instructions on how to modify your docker command to use a non-release version of fmriprep.
I’ll submit a pull request to fmriprep, once it’s merged in, I can give instructions on how to modify your docker command to use a non-release version of fmriprep.