Summary of what happened:
I am a new user to MRIQC, and have been trying to get it to work on my BIDS-ified data. It consists of anatomical MRI scans only. For this particular subject, I had trouble running MRIQC in one go. When I removed the problematic files it ran perfectly. However, when I ran the problematic file isolated, MRIQC could also process it. So I am wondering as to why the MRIQC is unable to process all the files in one go.
I would very much appreciate any assistance as I have been stuck on this issue for a fairly long time now, thank you in advance!
Command used (and if a helper script was used, a link to the helper script or the command generated):
docker run -it --rm -v /media/angkm/DataHDD/vascularDementia/mriqc_test/mriqc_test_dcm2bids_sub-01:/data:ro -v /media/angkm/DataHDD/vascularDementia/mriqc_test/mriqc_test_dcm2bids_sub-01/derivatives/mriqc:/out nipreps/mriqc:latest /data /out participant --participant_label 01 --no-sub --verbose-reports
Version:
23.1.0
Environment (Docker, Singularity, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
Relevant log outputs (up to 20 lines):
Traceback (most recent call last):
File "/opt/conda/bin/mriqc", line 8, in <module>
sys.exit(main())
File "/opt/conda/lib/python3.9/site-packages/mriqc/cli/run.py", line 168, in main
mriqc_wf.run(**_plugin)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/workflows.py", line 638, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/opt/conda/lib/python3.9/site-packages/mriqc/engine/plugin.py", line 184, in run
self._clean_queue(jobid, graph, result=result)
File "/opt/conda/lib/python3.9/site-packages/mriqc/engine/plugin.py", line 256, in _clean_queue
raise RuntimeError("".join(result["traceback"]))
RuntimeError: Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/mriqc/engine/plugin.py", line 60, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node measures.
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/mriqc/interfaces/anatomical.py", line 114, in _run_interface
raise RuntimeError(
RuntimeError: Input inhomogeneity-corrected data seem empty. MRIQC failed to process this dataset.
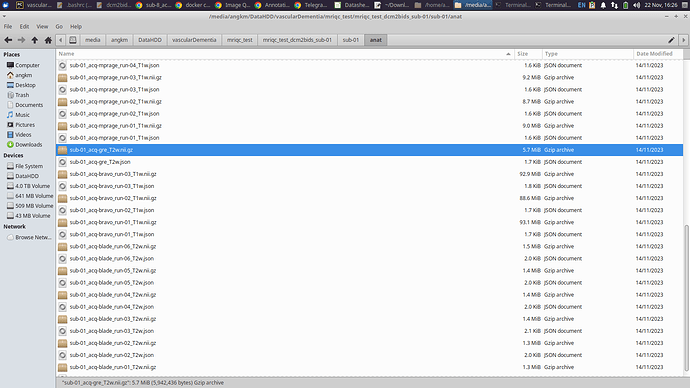
Screenshots / relevant information:
Below I have included some screenshots of the BIDS-ified subject that keeps encountering errors. For reference, the file sub-01_acq-gre_T2w.nii.gz is one of the problematic file.