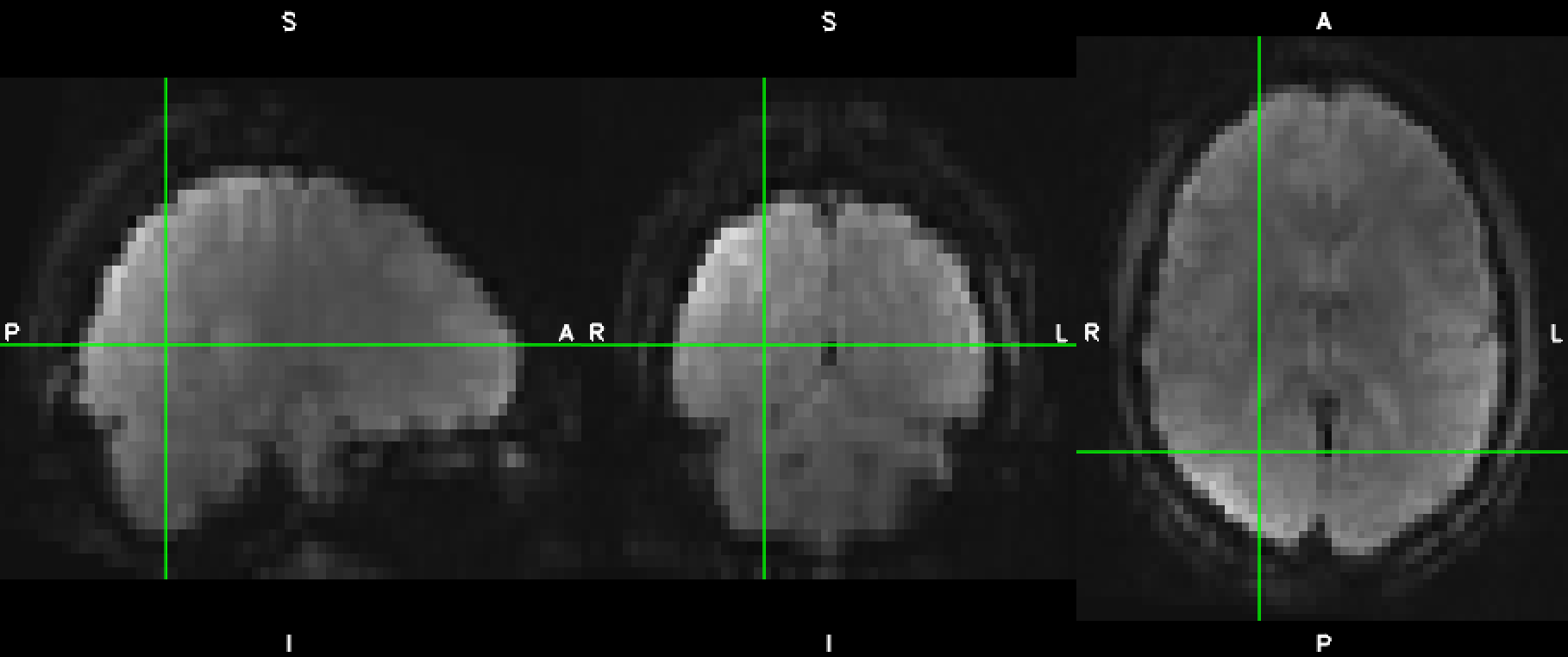
While I can see that a brain_mask file has been created, I can still see the skull in my preproc_bold results. I’ve attached sub-11_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz:
Should I be using a different skull-stripping template?
Also, I would like for my results to have different dimensions. Right now, they are 60x71x40:
Ideally, I’d like them to be either 91x109x91 with a voxel size of 2x2x2 or 60x71x60 with a voxel size of 3x3x3. Would a different template work? I’m just using the default at the moment.
This is the command I’m running:
singularity run jpetucci-fmriprep_icsaci-master-rec.simg anesthesia_dataset anesthesia_results/11/results participant --participant-label 11 --fs-license-file license.txt --mem_mb 10000 --force-bbr -w anesthesia_results/11/reports
I am running v1.4.1rc1.
Any help would be appreciated. Thank you!
Hi @rosaleen, thanks for the question.
The desc-preproc_bold.nii.gz files are not skull-stripped, but we provide a mask desc-brain_mask.nii.gz files that are in the same space. This is generally useful for assessing the quality of the mask. Because many tools allow you to load a BOLD series and a mask simultaneously, it seemed wasteful to output a second time series with the mask applied. However, if you need it, you can get a skull-stripped time series with fslmaths:
fslmaths sub-11_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz \
-mas sub-11_task-rest_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz \
out_file.nii.gz
You should be able to do this with --output-spaces MNI152NLin2009cAsym_desc:res-2. You can add other spaces, as well, if desired. See the Workflow configuration and the Spaces documentation.
3 Likes
I tried this:
singularity run jpetucci-fmriprep_icsaci-master-rec.simg
anesthesia_dataset
anesthesia_results/11/results
participant --participant-label 11
–fs-license-file license.txt
–mem_mb 10000
–fs-no-reconall
–force-bbr
–output-spaces MNI152NLin2009cAsym_desc:res-2
-w anesthesia_results/11/reports
and it said that
‘This is embarrassing - custom templates are not (yet) supported.’
NotImplementedError: This is embarrassing - custom templates are not (yet) supported.Please make sure none of the options already available via TemplateFlow fit your needs.
Do I need to update fmriprep?
I’m sorry, I made a copy-paste error. It should have been --output-spaces MNI152NLin2009cAsym:res-2. That _desc snuck in there.
1 Like