Summary of what happened:
I am trying to rerun some analysis using ALFF images from two cohorts, and I am having a hard time generating the actual ALFF images. A former member of my lab was able to successfully create these images, and I am using the same commands that they used, so I’m not really sure where I’m going wrong.
The images have been preprocessed using fmriprep. Everything else appears to be generating successfully, and there’s no clear error displaying in the log messages.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv \
--no-home \
-B ${jobTmpDir}:/tmp \
-B "/appl/freesurfer-7.1.1:/freesurfer" \
-B "/project/oathes_analysis2/individual_projects/julie/scripts/alff_project/xcp_d:/bidsfilt" \
-B "/project/oathes_analysis2/templateflow:/templateflow" \
-B ${input_dir}:/data/input \
-B ${output_dir}:/data/output \
/project/oathes_analysis2/singularity_containers/xcp_d-0.5.0.simg \
/data/input /data/output participant --participant_label $1 \
--work_dir ${SINGULARITYENV_TMPDIR} \
--bids-filter-file /bidsfilt/bids_filter_file.json \
--fs-license-file /freesurfer/license.txt \
--nthreads $2 \
--omp_nthreads $3 \
-vvv \
--lower-bpf 0.01 \
--upper-bpf 0.08 \
--input-type fmriprep \
--smoothing 4 \
-p 36P \
--despike
Version:
xcp_d-0.5.0.simg
Environment (Docker, Singularity / Apptainer, custom installation):
Singulary, submitting to LPC computing cluster
Relevant log outputs (up to 20 lines):
Crash output for a few of the subjects reflects an issue in the atlas space processing, but nothing about ALFF? Unless the ALFF calculation relies on atlas space workflow completing correctly.
cat crash-20250110-150701-dickson4-ds_atlas-b1fba21f-0dcb-4df9-9cac-414980f61864.txt
Error: Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 453, in aggregate_outputs
setattr(outputs, key, val)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/traits_extension.py", line 424, in validate
value = super(MultiObject, self).validate(objekt, name, newvalue)
File "/usr/local/miniconda/lib/python3.8/site-packages/traits/trait_types.py", line 2515, in validate
return TraitListObject(self, object, name, value)
File "/usr/local/miniconda/lib/python3.8/site-packages/traits/trait_list_object.py", line 582, in __init__
super().__init__(
File "/usr/local/miniconda/lib/python3.8/site-packages/traits/trait_list_object.py", line 213, in __init__
super().__init__(self.item_validator(item) for item in iterable)
File "/usr/local/miniconda/lib/python3.8/site-packages/traits/trait_list_object.py", line 213, in <genexpr>
super().__init__(self.item_validator(item) for item in iterable)
File "/usr/local/miniconda/lib/python3.8/site-packages/traits/trait_list_object.py", line 865, in _item_validator
return trait_validator(object, self.name, value)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/usr/local/miniconda/lib/python3.8/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: Each element of the 'out_file' trait of a _DerivativesDataSinkOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/data/output/xcp_d/space-MNI152NLin6Asym_atlas-Schaefer617_res-2_dseg.nii.gz' <class 'str'> was specified.
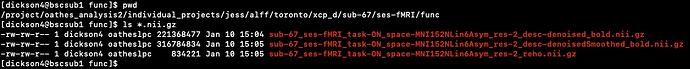
Screenshots / relevant information:
I’m expecting the *alff.nii.gz image to appear here as it did for the older runs.
I’m happy to provide any additional information as needed. Thanks in advance!