I downloaded the hcp data set and want to run a first-level analysis using SPM on it (to create beta-images). I know that I theoretically can only rely on the /tmp files that are created during the workflow execution (which are deleted after the workflow ran), however I would like to tweak the pipeline in such a way that the SPM.mat file and the corresponding images are saved locally and that other users who don’t want to use nipype can simply run the estimation using their SPM bare-metal installation, the respective NIFTI files and the SPM.mat file. For this, all my file paths have to point to paths on my local machine and not to the temporarily saved tmp/ files. Is there any way how to save the SPM.mat file in such a way that all its settings point to my output file:
/output/952863/first_level_pipe_nback/952863.nii?
Here’s my data structure (only one dummy subject). People that have experience with the HCP dataset should be familiar with this (I show only the showing the folders, not the files):
\DATA
└───952863
├───.xdlm
│ 952863_3T_tfMRI_WM_preproc.json
│
├───MNINonLinear
│ └───Results
│ ├───tfMRI_WM
│ │ prepare_level2_feat_analysis.sh
│ │ tfMRI_WM_hp200_s4_level2.fsf
│ │
│ ├───tfMRI_WM_LR
│ │ │ brainmask_fs.2.nii.gz
│ │ │ Movement_AbsoluteRMS.txt
│ │ │ Movement_AbsoluteRMS_mean.txt
│ │ │ Movement_Regressors.txt
│ │ │ Movement_Regressors_dt.txt
│ │ │ Movement_RelativeRMS.txt
│ │ │ Movement_RelativeRMS_mean.txt
│ │ │ PhaseOne_gdc_dc.nii.gz
│ │ │ PhaseTwo_gdc_dc.nii.gz
│ │ │ REC_run2_TAB.txt
│ │ │ SBRef_dc.nii.gz
│ │ │ tfMRI_WM_LR.L.native.func.gii
│ │ │ tfMRI_WM_LR.nii.gz
│ │ │ tfMRI_WM_LR.R.native.func.gii
│ │ │ tfMRI_WM_LR_Atlas.dtseries.nii
│ │ │ tfMRI_WM_LR_Atlas_MSMAll.dtseries.nii
│ │ │ tfMRI_WM_LR_hp200_s4_level1.fsf
│ │ │ tfMRI_WM_LR_Jacobian.nii.gz
│ │ │ tfMRI_WM_LR_Physio_log.txt
│ │ │ tfMRI_WM_LR_SBRef.nii.gz
│ │ │ WM_run2_TAB.txt
│ │ │
│ │ ├───EVs
│ │ │ 0bk_body.txt
│ │ │ 0bk_cor.txt
│ │ │ 0bk_err.txt
│ │ │ 0bk_faces.txt
│ │ │ 0bk_nlr.txt
│ │ │ 0bk_places.txt
│ │ │ 0bk_tools.txt
│ │ │ 2bk_body.txt
│ │ │ 2bk_cor.txt
│ │ │ 2bk_err.txt
│ │ │ 2bk_faces.txt
│ │ │ 2bk_nlr.txt
│ │ │ 2bk_places.txt
│ │ │ 2bk_tools.txt
│ │ │ all_bk_cor.txt
│ │ │ all_bk_err.txt
│ │ │ Sync.txt
│ │ │ WM_Stats.csv
│ │ │
│ │ └───RibbonVolumeToSurfaceMapping
│ │ goodvoxels.nii.gz
│ │
│ └───tfMRI_WM_RL
│ │ brainmask_fs.2.nii.gz
│ │ Movement_AbsoluteRMS.txt
│ │ Movement_AbsoluteRMS_mean.txt
│ │ Movement_Regressors.txt
│ │ Movement_Regressors_dt.txt
│ │ Movement_RelativeRMS.txt
│ │ Movement_RelativeRMS_mean.txt
│ │ PhaseOne_gdc_dc.nii.gz
│ │ PhaseTwo_gdc_dc.nii.gz
│ │ REC_run1_TAB.txt
│ │ SBRef_dc.nii.gz
│ │ tfMRI_WM_RL.L.native.func.gii
│ │ tfMRI_WM_RL.nii.gz
│ │ tfMRI_WM_RL.R.native.func.gii
│ │ tfMRI_WM_RL_Atlas.dtseries.nii
│ │ tfMRI_WM_RL_Atlas_MSMAll.dtseries.nii
│ │ tfMRI_WM_RL_hp200_s4_level1.fsf
│ │ tfMRI_WM_RL_Jacobian.nii.gz
│ │ tfMRI_WM_RL_Physio_log.txt
│ │ tfMRI_WM_RL_SBRef.nii.gz
│ │ WM_run1_TAB.txt
│ │
│ ├───EVs
│ │ 0bk_body.txt
│ │ 0bk_cor.txt
│ │ 0bk_err.txt
│ │ 0bk_faces.txt
│ │ 0bk_nlr.txt
│ │ 0bk_places.txt
│ │ 0bk_tools.txt
│ │ 2bk_body.txt
│ │ 2bk_cor.txt
│ │ 2bk_err.txt
│ │ 2bk_faces.txt
│ │ 2bk_nlr.txt
│ │ 2bk_places.txt
│ │ 2bk_tools.txt
│ │ all_bk_cor.txt
│ │ all_bk_err.txt
│ │ Sync.txt
│ │ WM_Stats.csv
│ │
│ └───RibbonVolumeToSurfaceMapping
│ goodvoxels.nii.gz
│
├───release-notes
│ tfMRI_WM_preproc.txt
│
└───T1w
└───Results
├───tfMRI_WM_LR
│ PhaseOne_gdc_dc.nii.gz
│ PhaseTwo_gdc_dc.nii.gz
│ SBRef_dc.nii.gz
│
└───tfMRI_WM_RL
PhaseOne_gdc_dc.nii.gz
PhaseTwo_gdc_dc.nii.gz
SBRef_dc.nii.gz
And here’s the code I have so far (strongly adapted from Michael Notter’s nipype-tutorial, and mostly dummy code that only serves the purpose of explaining the problem):
Code
import pandas as pd
from nipype.interfaces.utility import Rename
from nipype.interfaces.base import Bunch
from nipype.interfaces.io import SelectFiles, DataSink
from nipype import Workflow,Node,MapNode
from nipype.interfaces.utility import Function, IdentityInterface
from nipype.algorithms.misc import Gunzip
from os.path import join as opj
from nipype.interfaces.spm import Level1Design, EstimateModel, EstimateContrast
from nipype.algorithms.modelgen import SpecifySPMModel
# set data directory
data_dir = '/data/'
# take only this one subject
subject_list = ['952863']
# create a node that iterates over the subject(s)
subject_iterator = Node(IdentityInterface(fields=["subject_id"]),name="subject_iterator")
subject_iterator.iterables = [("subject_id",subject_list)]
# create a node that selects only the LR-n-back NIFTI file based on a template
templates = {'func':'{subject_id}/MNINonLinear/Results/tfMRI_WM_LR/tfMRI_WM_LR.nii.gz'}
selectfiles = Node(SelectFiles(templates,base_directory=data_dir),name='selectfiles')
# create a function that graps the event files from a run and converts them to a Bunch object
# NOTE! We have to import Bunch and pandas again because in nipype functions are closed environments
def get_nback_task_info(subject_id):
import pandas as pd
from nipype.interfaces.base import Bunch
# create a path to the directory where the event files are stored based on the input subject id
subject_dir = f"/data/{subject_id}/MNINonLinear/Results/tfMRI_WM_LR/EVs/"
# set up the names for all different event files
ev_txt_list = ['0bk_faces.txt','0bk_places.txt','0bk_body.txt','0bk_tools.txt',
'2bk_faces.txt','2bk_places.txt','2bk_body.txt','2bk_tools.txt']
# provide a list of names for all different event files
ev_names = ['0bk_faces','0bk_places','0bk_body','0bk_tools',
'2bk_faces','2bk_places','2bk_body','2bk_tools']
# initialize an empty list where the upcoming data frames will be stored
ev_dfs_list = []
for idx,ev_txt in enumerate(ev_txt_list):
# read in the .txt file as pandas data frame
ev_df = pd.read_table(subject_dir + ev_txt_list[idx],names=['onset','duration','amplitude'])
# add a column 'trial_type' to give a description
ev_df['trial_type'] = ev_names[idx]
# add df to event list
ev_dfs_list.append(ev_df)
# concatenate all dfs
ev_df = pd.concat(ev_dfs_list,axis=0)
# the ouput of our function will be passed over to SpecifyModel
# (https://nipype.readthedocs.io/en/1.6.0/api/generated/nipype.algorithms.modelgen.html#specifymodel)
# This class requires the input to be a Bunch-Object so we have to convert the dataframe to Bunch-Type
conditions = []
onsets = []
durations = []
for name,group in ev_df.groupby('trial_type'):
conditions.append(name)
onsets.append(group['onset'].tolist())
durations.append(group['duration'].tolist())
# the bunch object can contain more columns such as regressors (e.g. motion regressors?) that can be used to specfiy the model
nback_info = [Bunch(conditions=conditions,
onsets=onsets,
durations=durations,
#amplitudes=None,
#tmod=None,
#pmod=None,
#regressor_names=None,
#regressors=None
)]
return nback_info
# define a Node that takes the function above as input
nback_info = Node(Function(input_names=['subject_id'],
output_names=['nback_info'],
function=get_nback_task_info),
name='nback_info')
# create a node that unzips the original file (this is necessary for SPM to run)
gunzip = Node(Gunzip(),name='gunzip')
# create another node that dynamically changes the output filename based on the subject id
rename = Node(Rename(format_string="%(subject_id)s.nii"),name='rename_func_files')
# define a DataSink node where the (manipulated) files should be stored
# NOTE: The boolean parameterization = False ensures that the files are directly saved
# in the datasink folder. Otherwise for each subject there would be another folder _subject_123 created.
datasink = Node(DataSink(base_directory='/output',parameterization=False),name="datasink")
# Define SPM First Level Model
# SpecifyModel - Generates SPM-specific Model
modelspec = Node(SpecifySPMModel(concatenate_runs=False,
input_units='secs',
output_units='secs',
time_repetition=0.720,
high_pass_filter_cutoff=200),
name="modelspec")
# Level1Design - Generates an SPM design matrix
level1design = Node(Level1Design(bases={'hrf': {'derivs': [1, 0]}},
timing_units='secs',
interscan_interval=0.720,
model_serial_correlations='FAST'),
name="level1design")
# define workflow
wf = Workflow(name='spm_first_level_analysis_nback')
# pass the subject id over to selectfiles to select one file for that subject
wf.connect(subject_iterator,'subject_id',selectfiles,'subject_id')
# pass the subject id over to nback info to get the task information for that subject
wf.connect(subject_iterator,'subject_id',nback_info,'subject_id')
# pass the nback info to the modelspec node
wf.connect(nback_info,'nback_info',modelspec,'subject_info')
# pass the session info that is generated by SpecifySPM Model over to level-1 design node
wf.connect(modelspec,'session_info',level1design,'session_info')
# pass the file over to the Unzip Node to unzip it
# then pass the unzipped file over to the rename node
# also pass the subject id over to the rename node so that it knows how to rename the file using the subject_id
wf.connect(selectfiles,'func',gunzip,'in_file')
wf.connect(gunzip,'out_file',rename,'in_file')
wf.connect(subject_iterator,'subject_id',rename,'subject_id')
# pass the unzipped, renamed file over to modelspec
wf.connect(rename,'out_file',modelspec,'functional_runs')
# # Pass the unzipped, renamed file and the SPM.mat file to the datasink
# NOTE: Theoretically one doesn't even need to take this 'indirect' path from the datasink to the model
# you could also directly pass the file to the SPM model. But I need a way
# that other users who don't want to use nipype can run the SPM.mat files, so we are saving
# a hard copy of the files in each subject's directory. Also
wf.connect(subject_iterator,'subject_id',datasink,'container')
wf.connect(rename,'out_file',datasink,'first_level_pipe_nback.@unzipped_renamed_file')
wf.connect(level1design,'spm_mat_file',datasink,'first_level_pipe_nback.@spm_mat_file')
# Run the workflow
wf_results = wf.run()
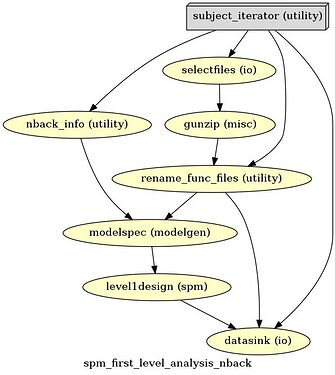
Here’s the output graph
# Create 1st-level analysis output graph
wf.write_graph(graph2use='colored', format='png', simple_form=True)
from IPython.display import Image
Image(filename='/home/neuro/nipype_tutorial/graph.png')
Have a look at the output
This looks like I want it to be.
\OUTPUT
└───952863
└───first_level_pipe_nback
952863.nii
SPM.mat
Problem
When having a look at the second node nback_info.a0, one can see why this can’t be run by SPM locally, because SPM of course does not have access to a file 'scans': '/tmp/tmpgim1n61i/spm_first_level_analysis_nback/_subject_id_955465/modelspec/955465.nii'that does not exist anymore:
node_list = list(wf_results.nodes())
node_list
[spm_first_level_analysis_nback.selectfiles.a0,
spm_first_level_analysis_nback.nback_info.a0,
spm_first_level_analysis_nback.modelspec.a0,
spm_first_level_analysis_nback.level1design.a0,
spm_first_level_analysis_nback.gunzip.a0,
spm_first_level_analysis_nback.rename_func_files.a0,
spm_first_level_analysis_nback.datasink.a0]
node_list[2].result.outputs
session_info = [{'cond': [{'name': '0bk_body', 'onset': [36.145], 'duration': [27.5]}, {'name': '0bk_faces', 'onset': [221.975], 'duration': [27.5]}, {'name': '0bk_places', 'onset': [250.257], 'duration': [27.5]}, {'name': '0bk_tools', 'onset': [107.48899999999999], 'duration': [27.5]}, {'name': '2bk_body', 'onset': [150.565], 'duration': [27.5]}, {'name': '2bk_faces', 'onset': [79.26100000000001], 'duration': [27.5]}, {'name': '2bk_places', 'onset': [178.7], 'duration': [27.5]}, {'name': '2bk_tools', 'onset': [7.997000000000001], 'duration': [27.5]}], 'hpf': 200.0, 'regress': [], 'scans': '/tmp/tmpgim1n61i/spm_first_level_analysis_nback/_subject_id_955465/modelspec/955465.nii'}]