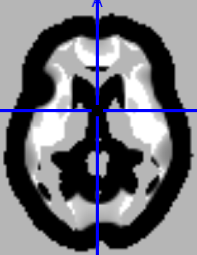
Summary of what happened:
I’m using the latest version of nilearn (0.10.4) but found that the results after estimating the 2nd level model (such as a one sample t-test) were suspiciously shrinked: the brain became smaller from each direction. It looks like an affine transform was applied. The output z map or p map still had the same image size as the input though. When I overlaid the z map into a standard brain, I could still see the brain outline but obviously the brain was squeezed from all direction and between the squeezed brain surface and the true outline, there is a band with equal thickness from all direction. The z value was -37. My input is individual nii.gz file but after I load the image, I did a z-transform and created a new nifti object using the affine and header from the corresponding image. I then concatenated the nifti object and input to the model when do fitting. This weird thing did not happen in previous nilearn.
Command used (and if a helper script was used, a link to the helper script or the command generated):
PASTE CODE HERE
here is the code
mv1 is either from the brain mask or one input image
niimgs = [nib.Nifti1Image(img, mv1.affine) for img in np.rollaxis(imgs, 3)]
design_matrix=pd.DataFrame( [1]*n, columns=[“intercept”] )
model = SecondLevelModel(smoothing_fwhm=8.0)
model=model.fit(niimgs, design_matrix=design_matrix)
resmaps = model.compute_contrast( second_level_contrast=“intercept”, output_type=“all”)
Version:
Environment (Docker, Singularity / Apptainer, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE
Screenshots / relevant information: