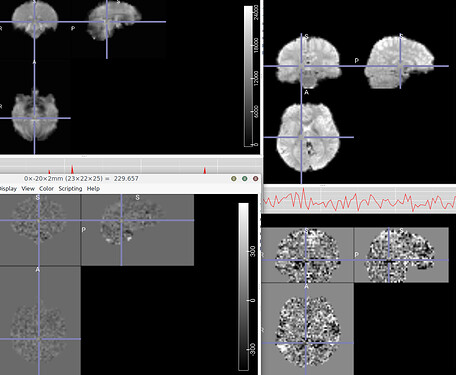
Hi all,
I’m a freshman in MRI data preprocessing. I perform filter (0.01-0.1) and detrend as follows, but my results seems problematic as attached (both in human and monkey), can anyone tell me why and how to fix this?
3dTproject -prefix filt.nii.gz -passband 0.01 0.1 -input rfMRI.nii.gz
3dTstat -mean -prefix filt_mean.nii.gz filt.nii.gz
3dDetrend -polort 2 -prefix dt.nii.gz filt.nii.gz
3dcalc -a filt_mean.nii.gz -b $dt.nii.gz -expr ‘a+b’ -prefix Final.nii.gz
The first row are original figures and the second are that filtered
Thank you so much
Jewel