Hi-
Just to comment on some features of AFNI.
Background, about spaces
-
In the NIFTI header, there are the sform_code and qform_code values, which are integers that essentially encode whether the dataset is in an “original” subject space or whethether it is in some “aligned” space, like a reference template. (Annoyingly, there is one code value, “2”, that is ambiguous, so it could be either—sigh. But let’s ignore that for the minute—only certain software populate that code value, and I don’t think it is being used here.) So, if you have a dataset with sform_code or qform_code 3, 4 or 5, that dataset is in a standard space, for example, namely Talairach-Tournous (TT), MNI or “other template”, respectively. You can use this to view the code values for one or more dsets:
nifti_tool -disp_hdr -field sform_code -field qform_code -infiles DSET1 [DSET2 DSET3 ...]
-
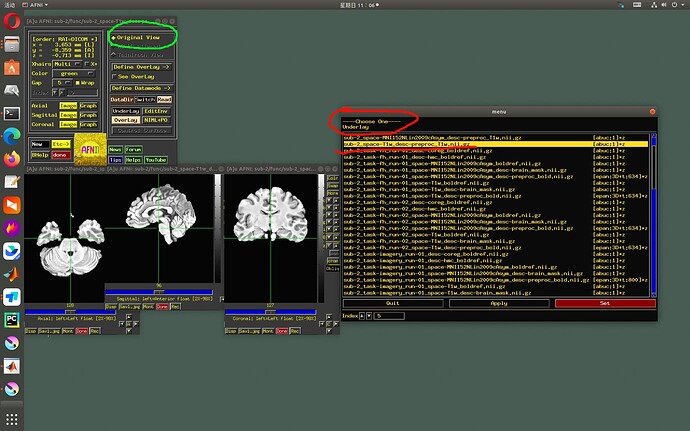
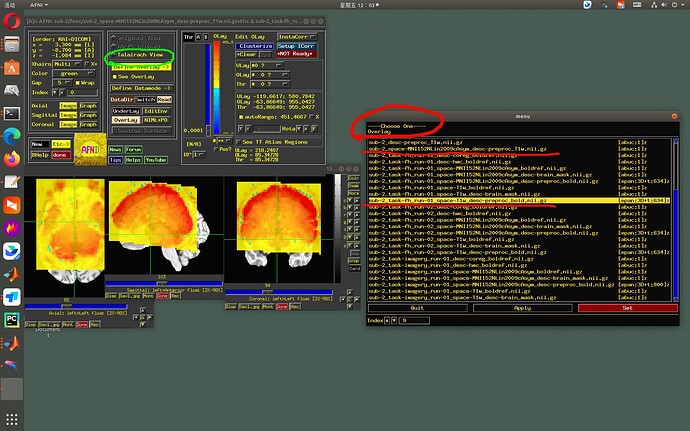
In AFNI, this concept is encoded in the “space” and “AFNI view space (av_space)” of the header, whether the dset itself is BRIK/HEAD or NIFTI format. If the dataset is in some original space, space will be “ORIG” and av_space will be “+orig”. If the dataset is in some standard space, the av_space will generically be “+tlrc” (NB: for historic reasons it is called this, but it now just reflects any template, not just TT); the “space” value will generally be more informative about the specific space name, such as “TT_N27”, “MNI”, “MNI_2009c_asym”, “HaskinsPeds”, “NMT2”, etc. To view the value of this for one or more dsets, whether the dataset is NIFTI or BRIK/HEAD, use:
3dinfo -space -av_space -prefix DSET1 [DSET2 DSET3 ...]
Some further notes on the sform_code and qform_code are here. Specific notes on NIFTI and BRIK/HEAD similarities and differences are here. Some notes on NIFTI (and specifically NIFTI+AFNI working together, since historically NIFTI has been primarily developed and maintained by AFNI developers) are [here](8.1. The NIFTI format — AFNI, SUMA and FATCAT: v24.0.02, though the ambiguous [qs]form_code values were included to maintain IMG/HDR format properties from other software.).
OK, now to the point of the GUI:
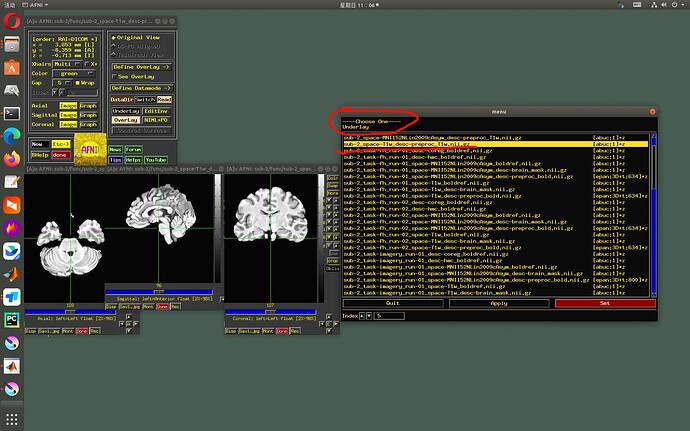
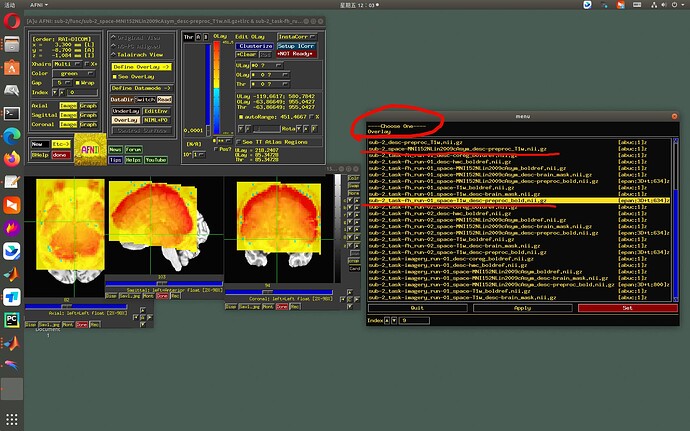
- An original design feature in the GUI was to prevent people from overlaying/underlaying datasets that were in a template space with ones in original space. The idea was that data in different spaces wouldn’t be expected to be aligned anyways, and that this might help prevent confusion among a myriad of datasets.
- Therefore, the AFNI GUI will not show an underlay-overlay pair together if one dset is in a standard space and the other is in a native space. That is, if:
- 3dinfo would show a “+tlrc” av_space for one and “+orig” space for the other
- NIFTI [sq]form_code value is 3, 4 or 5 for one dset and 0 or 1 for the other.
- Note that you can overlay any two templates on each other–so, you can overlay/underlay something with sform_code=4 with a dset having sform_code=5, or av_space “MNI” with av_space “NMT2”, say.
It would be odd to see a NIFTI dataset in subject T1w-space have a sform_code and qform_code that basically says, “hey, i’m in template space”. Is there some reason that that should happen? Is it possible that the sform_code/qform_code value of that dataset is the dreaded ambiguous “2”? Looking at a couple AFNI Message Board posts, I think that might be the case: see here.
So, if that is the case, you can direct AFNI how to treat the ambiguous qform_code and sform_code values of 2, using an AFNI environment variable. You can set this in your ~/.afnirc file of environment variables:
AFNI_NIFTI_VIEW = orig
After that, the ambiguously coded NIFTI datasets will be treated as having av_space “+orig” instead of “+tlrc”, which is how they must currently be treated. It would be nice to not have amiguous header information, though, if that were possible.
Re. obliquity: it is unfortunate to have obliquity in the anatomical dataset, often. Different software deal with it differently. Basically, the issue it causes is just a visualization one for any GUI:
- if you apply the obliquity, you have to re-grid or interpolate the data, and that creates some blurring, and you can’t click on a location and see the actual value from the real dataset.
- if you ignore the obliquity*, you click somewhere on the brain and see the correct, exact, uninterpolated values in the data, but the (x,y,z) coordinate location there is not what it was in the scanner, and if you try to overlay/underlay another dataset that has a different grid and/or obliquity, they won’t overlap properly.
Different softwares make different choices for visualizing. Some apply the obliquity automatically (I think FSL does this?), and AFNI chooses to ignore the obliquity part. Either way, it’s a tradeoff, but you should know what is going to happen, certainly.
During processing, obliquity should just be dealt with by the tools properly, so again the visualization considerations are separate (though visualizing data is important!).
Having EPIs with obliquity is fine, because by the end of preprocessing they generally have been aligned to something else and you don’t have to worry about obliquity considerations anymore. For anatomical dsets to have obliquity can be a bit more painful, as you are seeing. We typically recommend that people remove the obliquity from those before preprocessing, to not cause issues/mismatched assumptions, esp. across software. Typically, anatomical obliquity is not too large—if it is, or if one is doing specialized slab/slice alignment, one might want to not do this, but except in those cases, you could consider getting rid of obliquity in a way that neither interpolates the data nor changes the coordinate origin (x,y,z)=(0,0,0) by doing this for your DSET_IN (and see this thread):
3dcopy DSET_IN _tmp_dset # copy to BRIK/HEAD
3drefit -oblique_recenter _tmp_dset+orig # recenter, obl transform is now just mainly rotation
3drefit -deoblique _tmp_dset+orig # purge obl info
# ... and then if you want a NIFTI output
3dcopy _tmp_dset+orig DSET_OUT.nii
Another option would be to just apply the obliquity of the anatomical dataset, using 3dWarpDrive. We did that in processing the NARPS data, for example:
3dWarp \
-deoblique \
-wsinc5 \
-prefix ${sdir_deob}/${subj}_T1w-deobl.nii.gz \
-gridset ${grid_template} \
${dset_anat_raw}
That incurs a slight blurring during the regridding, which using -wsinc5 minimizes. This is the approach that FreeSurfer’s recon-all would take if you input an oblique anatomical: apply the obliquity and process from there.
Sorry for the long post here, but there were several topics to cover. I hope some of that is useful.
–pt