Summary of what happened:
Hello Neurostars community!
I am posting to this community because I have been trying to post-process HCP-D data with the XCP-D pipeline to obtain ALFF measures for the last few weeks. Specifically, I have been trying to replicate the patterns observed in (Intrinsic activity development unfolds along a sensorimotor–association cortical axis in youth | Nature Neuroscience), but I have observed quite different spatial patterns, and I am pretty sure that something has gone wrong in my pipeline (since these patterns have been robustly observed in multiple datasets).
Setup:
- Data: HCP-D dataset, downloaded with Datalad
- Container: Singularity container for XCP-D
- Version: XCP-D v0.10.6 (i.e., latest stable version)
Problem:
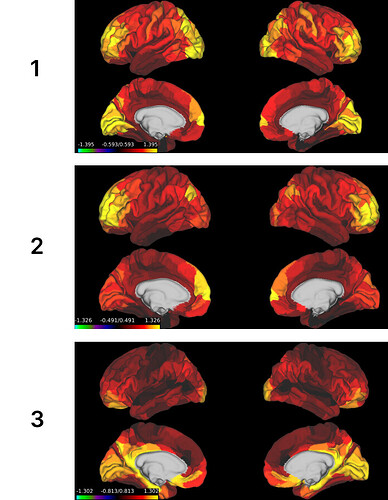
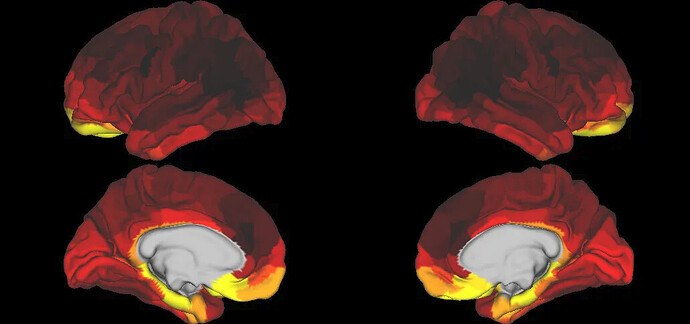
When comparing the ALFF values averaged across subjects, the spatial patterns for the ALFF look very different from other datasets (which seem more consistent across datasets) and also from the patterns I have seen for the youngest 100 HCP subjects (see images at the end of this post).
I ran the pipeline with the following command (I did not specify all parameters as I was using linc mode and therefore used the default parameters):
Command used:
singularity run -B /tmp/xcp_input_${subject} --cleanenv /data/project/sh_dev/xcp_d.sif /tmp/xcp_input_${subject} /tmp/xcp_output_${subject} participant \
--despike --lower-bpf 0.01 --upper-bpf 0.08 -p 36P -f 10 --input_type hcp --mode linc --fd-thresh 0
I have used the uncleaned data as it was listed in the ingression module for the required HCP derivatives (see: xcp_d.ingression.hcpya module — xcp_d 0.10.8.dev100+gc3c811b documentation). However, I renamed the HCP-D Data as the acquisition was anterior/posterior compared to left/right as in HCP-YA and the bids convert function required LR/RL naming to identify files.
These are the specifications from the toml file of a single subject:
[workflow]
mode = "linc"
file_format = "cifti"
dummy_scans = 0
input_type = "hcp"
despike = true
smoothing = 6
output_interpolated = false
combine_runs = false
motion_filter_order = 4
head_radius = 50
fd_thresh = 0.0
min_time = 0
bandpass_filter = true
high_pass = 0.01
low_pass = 0.08
bpf_order = 2
min_coverage = 0.5
correlation_lengths = [ "all",]
process_surfaces = false
abcc_qc = false
linc_qc = true
[execution]
fmri_dir = "/tmp/working_dir/dset_bids/derivatives/hcp"
aggr_ses_reports = 4
bids_database_dir = "/tmp/working_dir/20250326-120925_7415aed1-785a-4e92-872d-3ec5ccecef91/bids_db"
bids_description_hash = "fd723227bf2c3d9b08e6f41318d87c925868a516aebc647cbd2b6556f9d036f7"
boilerplate_only = false
confounds_config = "/usr/local/miniconda/lib/python3.10/site-packages/xcp_d/data/nuisance/36P.yml"
debug = []
layout = "BIDS Layout: ..._dir/dset_bids/derivatives/hcp | Subjects: 1 | Sessions: 0 | Runs: 2"
log_dir = "/tmp/xcp_output_HCD0001305_V1_MR/logs"
log_level = 25
low_mem = false
md_only_boilerplate = false
notrack = false
reports_only = false
output_dir = "/tmp/xcp_output_HCD0001305_V1_MR"
atlases = [ "4S1056Parcels", "4S156Parcels", "4S256Parcels", "4S356Parcels", "4S456Parcels", "4S556Parcels", "4S656Parcels", "4S756Parcels", "4S856Parcels", "4S956Parcels", "Glasser", "Gordon", "HCP", "MIDB", "MyersLabonte", "Tian",]
run_uuid = "20250326-120925_7415aed1-785a-4e92-872d-3ec5ccecef91"
participant_label = [ "HCD0001305",]
templateflow_home = "/home/sobotta/.cache/templateflow"
work_dir = "/tmp/working_dir"
write_graph = false
Things I have noticed that might be problematic:
- The only error running the pipeline was the linc quality control - it did not work because it failed to identify the relevant files, see:
Node inputs:
TR = 0.8
anat_mask_anatspace = <undefined>
bold_file = <undefined>
bold_mask_anatspace = <undefined>
bold_mask_inputspace = None
bold_mask_stdspace = <undefined>
cleaned_file = <undefined>
dummy_scans = <undefined>
head_radius = 50.0
motion_file = <undefined>
name_source = /data/project/sh_dev/work/dset_bids/derivatives/hcp/sub-HCD0029630_V1_MR/func/sub-HCD0029630_V1_MR_task-rest_dir-LR_run-1_space-fsLR_den-91k_bold.dtseries.nii
template_mask = /home/sobotta/.cache/templateflow/tpl-MNI152NLin2009cAsym/tpl-MNI152NLin2009cAsym_res-02_desc-brain_mask.nii.gz
temporal_mask = <undefined>
- In the BIDS layout it lists two runs and zero sessions even though there are four runs (two for each session), however, all four runs have been processed and are also different from each other.
- The values I obtain from the xcp-d output for ALFF are quite large in scale. Even though this can be fixed by mean scaling, I was wondering whether this is already an indication of an issue.
- I am only using the MSMAll-aligned functional data as this is what was specified in the HCP-YA example. Maybe it could be beneficial to also use MSMAll-aligned structural data?
These are some of the things I have tried:
- calculated ALFF manually for comparison (with two different implementations) based on HCP denoised data (i.e., HCP minimally preprocessed with ICA-fix denoising): I saw very similar patterns.
- different parcellations, nuissance regressors, etc.
- run the xcp-d with different parameters (also including motion scrubbing):
singularity run -B /data/project/sh_dev/data/singlesub_HCD0001305_V1_MR --cleanenv /data/project/sh_dev/xcp_d.sif /data/project/sh_dev/data/singlesub_HCD0001305_V1_MR /data/project/sh_dev/data/singlesub_outputBianca36P_HCD0001305_V1_MR participant \
--despike y --lower-bpf 0.01 --upper-bpf 0.08 --input_type hcp --mode none --fd-thresh 0.3 --combine-runs n --atlases Glasser --linc-qc y --warp-surfaces-native2std n --smoothing 6 --file-format cifti --dummy-scans auto --abcc-qc n \
--min-coverage 0.5 --motion-filter-type none --output-type censored --nuisance-regressors 36P --head-radius 50 --min-time 240 --create-matrices 240 all --work-dir /data/project/sh_dev/work
If someone in this community has an idea what could be the reason for this spatial patterns or spots a potential problem in the postprocessing, that would be super helpful. Any help, comments or input are warmly welcomed. Thank you very much in advance for your time!
Best wishes,
Sarah
Screenshots / relevant information:
- Average ALFF values for HCP 100 youngest subjects (obtained from Valerie Sydnor)
- Average ALFF values for PNC 100 oldest subjects (obtained from Valerie Sydnor):
- My average ALFF values based on a sample of 464 subjects from HCP-D (8-21 years).
The first two show great consistency across ages and data sets, and we can observe this shift to larger values in the PFC and reductions in visual areas with increasing age.
However, my ALFF values show some similarity in spatial patterns, but also large differences, with the greatest activation in the isthmus cingulate and areas.
Additionally, I also uploaded the html files for a sample subject run with the two different xcp-d parameters, in case this is helpful: ALFF – Google Drive