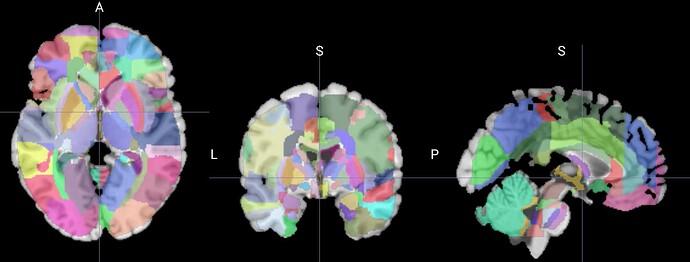
What should I do to have the JHU atlas matching the border of the brain in the CH2 template.
As you can see below there is a mismatch.
I tried a couple of things with flirt and nibabel but I failed each time.
Hi @simon.thibault,
You would need to find T1w images corresponding to both of the templates, then compute a registration between one and the other. Then apply that registration to the label image using nearest neighbor (or genericLabel if using ANTs) interpolation, keeping in mind that the same template space you had as the moving image for the registration should be the label image that is moved for the warping step.
Best,
Steven
Thanks, I don’t think I could find the corresponding T1w for the atlas image. This image is provided without further detail in mricron. After further searches I think I found a more recent images for this atlas and this is registered to the same template space. GitHub - muschellij2/Eve_Atlas: Eve Atlas used for segmentations
Thanks,
Simon