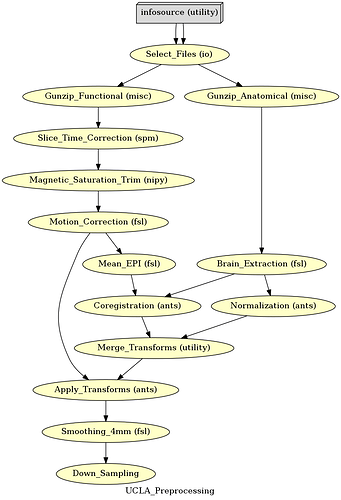
I’m performing Preprocessing using Nipype and am pretty new to the neuroimaging domain. My preprocessing workflow is executing fine but the dimension and size are increasing drastically after the ApplyTransform from ANTs.
This is my Workflow:
These are the parameters that I have for Registration, Normalization and Transformation
Coregistration
from nipype.interfaces.ants import Registration
coregistration = Node(Registration(), name='Coregistration')
coregistration.inputs.float = False
coregistration.inputs.output_transform_prefix = 'meanEpi2highres'
coregistration.inputs.transforms = ['Rigid']
coregistration.inputs.transform_parameters = [(0.1,), (0.1,)]
coregistration.inputs.number_of_iterations = [[1000, 500, 250, 100]]
coregistration.inputs.dimension = 3
coregistration.inputs.num_threads = THREADS
coregistration.inputs.write_composite_transform = True
coregistration.inputs.collapse_output_transforms = True
coregistration.inputs.metric = ['MI']
coregistration.inputs.metric_weight = [1]
coregistration.inputs.radius_or_number_of_bins = [32]
coregistration.inputs.sampling_strategy = ['Regular']
coregistration.inputs.sampling_percentage = [0.25]
coregistration.inputs.convergence_threshold = [1e-08]
coregistration.inputs.convergence_window_size = [10]
coregistration.inputs.smoothing_sigmas = [[3, 2, 1, 0]]
coregistration.inputs.sigma_units = ['mm']
coregistration.inputs.shrink_factors = [[4, 3, 2, 1]]
coregistration.inputs.use_estimate_learning_rate_once = [True]
coregistration.inputs.use_histogram_matching = [False]
coregistration.inputs.initial_moving_transform_com = True
coregistration.inputs.output_warped_image = True
coregistration.inputs.winsorize_lower_quantile = 0.01
coregistration.inputs.winsorize_upper_quantile = 0.99
Normalization
normalization = Node(Registration(), name='Normalization')
normalization.inputs.float = False
normalization.inputs.collapse_output_transforms = True
normalization.inputs.convergence_threshold = [1e-06]
normalization.inputs.convergence_window_size = [10]
normalization.inputs.dimension = 3
normalization.inputs.fixed_image = MNItemplate
normalization.inputs.initial_moving_transform_com = True
normalization.inputs.metric = ['MI', 'MI', 'CC']
normalization.inputs.metric_weight = [1.0]*3
normalization.inputs.number_of_iterations = [[1000, 500, 250, 100],
[1000, 500, 250, 100],
[100, 70, 50, 20]]
normalization.inputs.num_threads = THREADS
normalization.inputs.output_transform_prefix = 'anat2template'
normalization.inputs.output_inverse_warped_image = True
normalization.inputs.output_warped_image = True
normalization.inputs.radius_or_number_of_bins = [32, 32, 4]
normalization.inputs.sampling_percentage = [0.25, 0.25, 1]
normalization.inputs.sampling_strategy = ['Regular', 'Regular', 'None']
normalization.inputs.shrink_factors = [[8, 4, 2, 1]]*3
normalization.inputs.sigma_units = ['vox']*3
normalization.inputs.smoothing_sigmas = [[3, 2, 1, 0]]*3
normalization.inputs.transforms = ['Rigid', 'Affine', 'SyN']
normalization.inputs.transform_parameters = [(0.1,), (0.1,), (0.1, 3.0, 0.0)]
normalization.inputs.use_histogram_matching = True
normalization.inputs.winsorize_lower_quantile = 0.005
normalization.inputs.winsorize_upper_quantile = 0.995
normalization.inputs.write_composite_transform = True
Apply Transform
from nipype.interfaces.utility import Merge
from nipype.interfaces.ants import Registration, ApplyTransforms
merge_transforms = Node(Merge(2), iterfield=['in2'], name='Merge_Transforms')
apply_transforms = Node(ApplyTransforms(), iterfield=['input_image'], name='Apply_Transforms')
apply_transforms.inputs.input_image_type = 3
apply_transforms.inputs.float = False
apply_transforms.inputs.num_threads = THREADS
apply_transforms.inputs.environ = {}
apply_transforms.inputs.interpolation = 'BSpline'
apply_transforms.inputs.invert_transform_flags = [False, False]
apply_transforms.inputs.reference_image = MNItemplate
The output of the Motion Control is (64, 64, 34, 147) around 24 MB
The output of Apply Transform is (91, 109, 91, 147) around 761 MB
The output of Brain Extraction is (176, 256, 256) around 2 MB
What’s the reason behind this and how do I resolve it?
Thanks in advance!