Summary of what happened:
Hi,
I am running a DTI analysis in FSL, but trying to use ANTs for better registration.
I am mainly focusing on the precuneus.
The precuneus mask is from the FSL’s Harvard - Oxford Atlas.
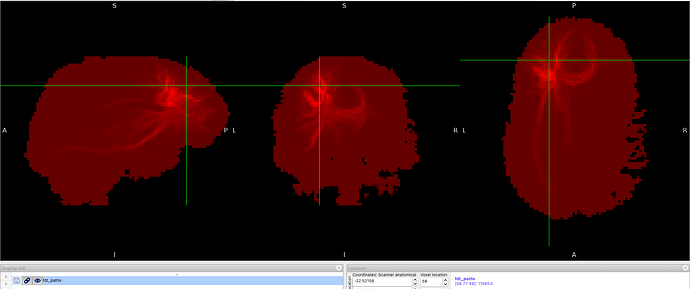
I run the registration in ANTs with the following code, however the output and the image I received was not quite the precuneus.
Can somebody assist me in the problem? Did I mix up the images at one of the registration steps?
This is my code:
Command used (and if a helper script was used, a link to the helper script or the command generated):
##00.0 ANTs-PATH
export PATH=/opt/ants/bin:$PATH
##00.1 defining & creating a folder
mkdir -p /Volumes/Diana/Preprocessed/DTI/Test_BedpostX/ANTs/T1_MNI
T1_folder="/Volumes/Diana/Preprocessed/DTI/Test_BedpostX/ANTs/T1_MNI"
##01.1 defining the path #to t1brain
template="/Users/anneliesvantwesteinde/fsl/data/standard/MNI152_T1_2mm_brain.nii.gz"
t1brain="/Volumes/Diana/Preprocessed/DTI/Test_BedpostX/ADDI_013/ADDI_013_T1/ADDI_013_T1.nii"
#01.2 Registration Atlas T1 - > subject T1 (non-linear):
antsRegistration -d 3 --float 0 --output "$T1_folder"/out \
--interpolation Linear \
--winsorize-image-intensities [0.005,0.995] \
--use-histogram-matching 0 \
--initial-moving-transform [$template,$t1brain,1] \
--transform Rigid[0.1] \
--metric MI[$template,$t1brain,1,32,Regular,0.25] \
--convergence [1000x500x250x100,1e-6,10] \
--shrink-factors 8x4x2x1 \
--smoothing-sigmas 3x2x1x0vox \
--transform Affine[0.1] \
--metric MI[$template,$t1brain,1,32,Regular,0.25] \
--convergence [1000x500x250x100,1e-6,10] \
--shrink-factors 8x4x2x1 \
--smoothing-sigmas 3x2x1x0vox \
--transform SyN[0.1,3,0] \
--metric CC[$template,$t1brain,1,4] \
--convergence [100x70x50x20,1e-6,10] \
--shrink-factors 8x4x2x1 \
--smoothing-sigmas 3x2x1x0vox \
--verbose 1
##02.1 defining the path
b0brain="/Volumes/Diana/Preprocessed/DTI/Addison_AllControls/ADDI_013/nodif.nii.gz"
t1brain_mask="/Volumes/Diana/Preprocessed/T1/BrainMasks/FSL_Anat_Addison_AllControls/ADDI_013_T1/ADDI_013_T1.anat/T1_biascorr_brain_mask.nii.gz"
#02.2creating a separate folder
mkdir -p "$T1_folder"/b0_T1
#02.3 Registration Subject b0 -> Subject T1 (linear/affine):
antsRegistrationSyN.sh -d 3 -f "$t1brain" -m "$b0brain" -o "$T1_folder/b0_T1/out" -n 4 -t a -x "$t1brain_mask"
##03.1 defining the path, creating another folder
ROI_mask="/Users/anneliesvantwesteinde/precuneus/precuneus_ROI_Left.nii.gz"
new_mask_dir="$T1_folder/"
mkdir -p "$T1_folder"/new_mask_dir
filename_mask=$(basename "$ROI_mask" .nii.gz)
#03.2 Then we bring the ROI from Atlas T1 -> subject T1 -> subject b0
antsApplyTransforms -d 3 -i "$ROI_mask" -r $b0brain -t ["$T1_folder"/b0_T1/out0GenericAffine.mat,1] -t ["$T1_folder"/out0GenericAffine.mat,1] -t "$T1_folder"/out1InverseWarp.nii.gz -n NearestNeighbor -o "$T1_folder/new_mask_dir/${filename_mask}_in_T2.nii.gz" -v
#04.01 defining
DWI_folder="/Volumes/Diana/Preprocessed/DTI/Test_BedpostX/ADDI_013.bedpostX"
directory_path="$DWI_folder/probtrackx"
mkdir -p "$directory_path"
#04. give exact path! "$filename_mask"_in_T2.nii.gz is the file you will supply to the probtrackx2. For example, like this:
probtrackx2 \
-x "/Volumes/Diana/Preprocessed/DTI/Test_BedpostX/ANTs/T1_MNI/new_mask_dir/precuneus_ROI_Left_in_T2.nii.gz" \
-V 1 \
-l \
--modeuler \
--onewaycondition -c 0.2 -S 2000 \
--steplength=0.5 \
-P 5000 \
--fibthresh=0.01 \
--distthresh=0.0 \
--sampvox=0.0 \
--forcedir \
--opd \
-s "$DWI_folder"/merged \
-m "$DWI_folder"/nodif_brain_mask.nii.gz \
--dir="$directory_path"
Version:
Version unknown ![]() - I downloaded and installed the ANTs a week ago, from github.
- I downloaded and installed the ANTs a week ago, from github.
Environment (Docker, Singularity / Apptainer, custom installation):
I am using bash for running this script and my operating system and version macOS-14.0; CPU architecture arm64 (Apple M1/M2)
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
Screenshots / relevant information:
Thank you in advance!