Hi all,
I’m having some problems normalising my data to MNI space using the ANTs Registration interface in nipype as part of the pipeline.
In short, the template image is warped to subj space using a binary mask of grey and white matter as the fixed image then we apply inverse transform to warp the subj image to MNI space. The data has lesion damage and we use a cost function lesion mask in the template > subj warp. Strangely this method has worked very well in the past.
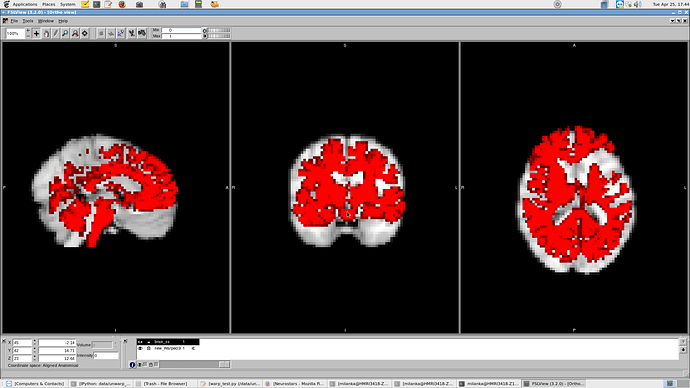
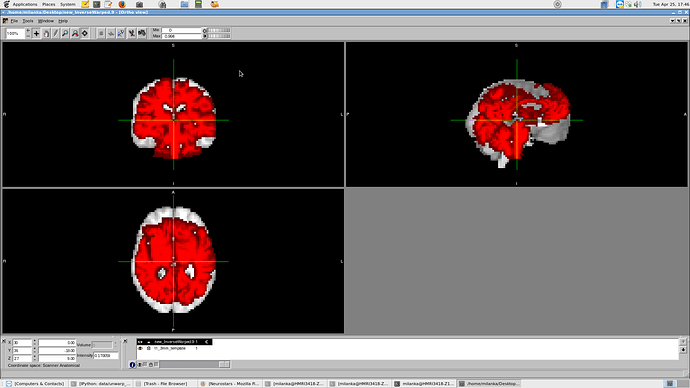
The template > subj space extends the frontal lobe too far (image 1, fixed image in red), while the subj > mni warp output warps for most (but not all) patients to be shrunk in the x and z and pitched too high in the y (image 2, inverse fixed image in red [see post 2]).
The only major difference between the dataset that this has worked previously to and the current is voxel sizes (3mm isotropic vs. 2.95 x 2.95 x 3).
The parameters are below.
antsnorm.inputs.output_transform_prefix = “new”
antsnorm.inputs.collapse_output_transforms=True
antsnorm.inputs.initial_moving_transform_com=True
antsnorm.inputs.num_threads=1
antsnorm.inputs.output_inverse_warped_image=True
antsnorm.inputs.output_warped_image=True
antsnorm.inputs.sigma_units=[‘vox’]*3
antsnorm.inputs.transforms=[‘Rigid’, ‘Affine’, ‘SyN’]
antsnorm.inputs.terminal_output=‘file’
antsnorm.inputs.winsorize_lower_quantile=0.005
antsnorm.inputs.winsorize_upper_quantile=0.995
antsnorm.inputs.convergence_threshold=[1e-06]
antsnorm.inputs.convergence_window_size=[10]
antsnorm.inputs.metric=[‘MI’, ‘MI’, ‘CC’]
antsnorm.inputs.metric_weight=[1.0]*3
antsnorm.inputs.number_of_iterations=[[1000, 500, 250, 100],[1000, 500, 250, 100],[100, 70, 50, 20]]
antsnorm.inputs.radius_or_number_of_bins=[32, 32, 4]
antsnorm.inputs.sampling_percentage=[0.25, 0.25, 1]
antsnorm.inputs.sampling_strategy=[‘Regular’,‘Regular’,‘None’]
antsnorm.inputs.shrink_factors=[[8, 4, 2, 1]]*3
antsnorm.inputs.smoothing_sigmas=[[3, 2, 1, 0]]*3
antsnorm.inputs.transform_parameters=[(0.1,),(0.1,),(0.1, 3.0, 0.0)]
antsnorm.inputs.use_histogram_matching=True
antsnorm.inputs.write_composite_transform=False
Any suggestions on where to start troubleshooting would be appreciated!
Thanks,
Peter