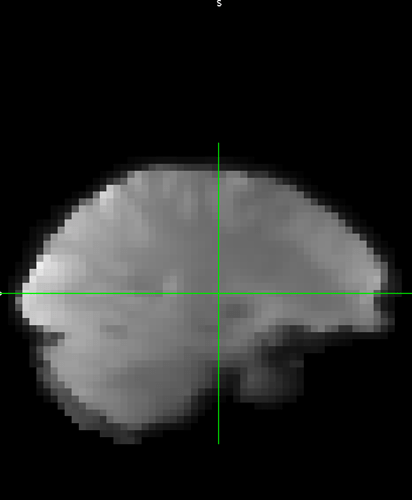
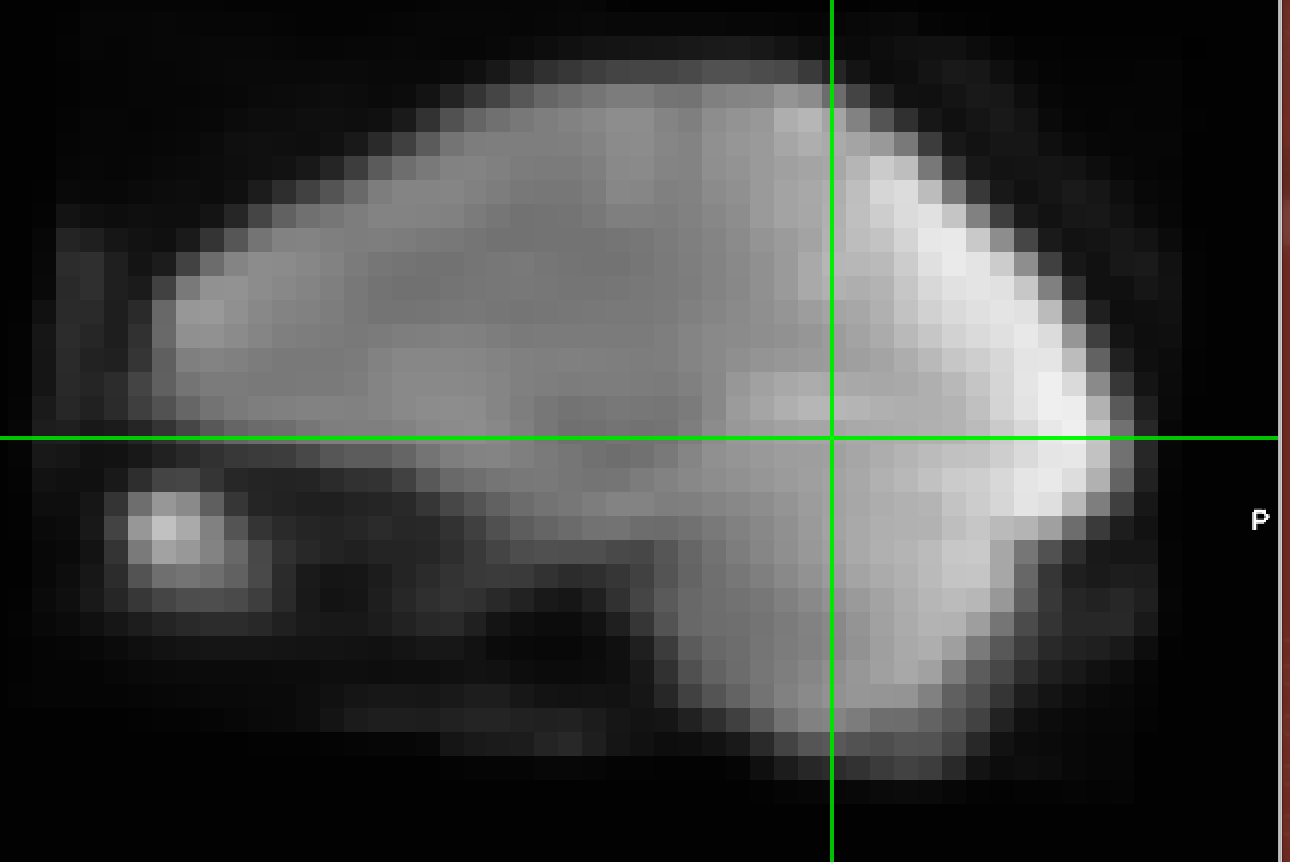
Hi everyone,
I’ve searched around and have not found anyone with this exact issue. I’ve been running a preprocessing pipeline based on miykael’s tutorial: https://miykael.github.io/nipype_tutorial/notebooks/example_preprocessing.html
While the code executes, my preprocessed images are coregistered to the original anatatomicals without the skull stripping and transformation. Extraction and segmentation both work as they should. However, despite what the code says somehow the non-extracted original image is being piped in to the ApplyWarp node. I’m not sure how the first line could pipe in the BET output image correctly to segmentation, but then this doesn’t happen int he 7th line. Here is the code:
coregwf.connect([(bet_anat, segmentation, [(‘out_file’, ‘in_files’)]),
(segmentation, threshold, [((‘partial_volume_files’, get_wm),
‘in_file’)]),
(bet_anat, coreg_pre, [(‘out_file’, ‘reference’)]),
(threshold, coreg_bbr, [(‘out_file’, ‘wm_seg’)]),# possibly where things are going wrong
(coreg_pre, coreg_bbr, [(‘out_matrix_file’, ‘in_matrix_file’)]),
(coreg_bbr, applywarp, [(‘out_matrix_file’, ‘in_matrix_file’)]),
(bet_anat, applywarp, [(‘out_file’, ‘reference’)]),
(coreg_bbr, applywarp_mean, [(‘out_matrix_file’, ‘in_matrix_file’)]),
(bet_anat, applywarp_mean, [(‘out_file’, ‘reference’)]),
])
Any help would be much appreciated. Please ask further details that you may need. Thanks!