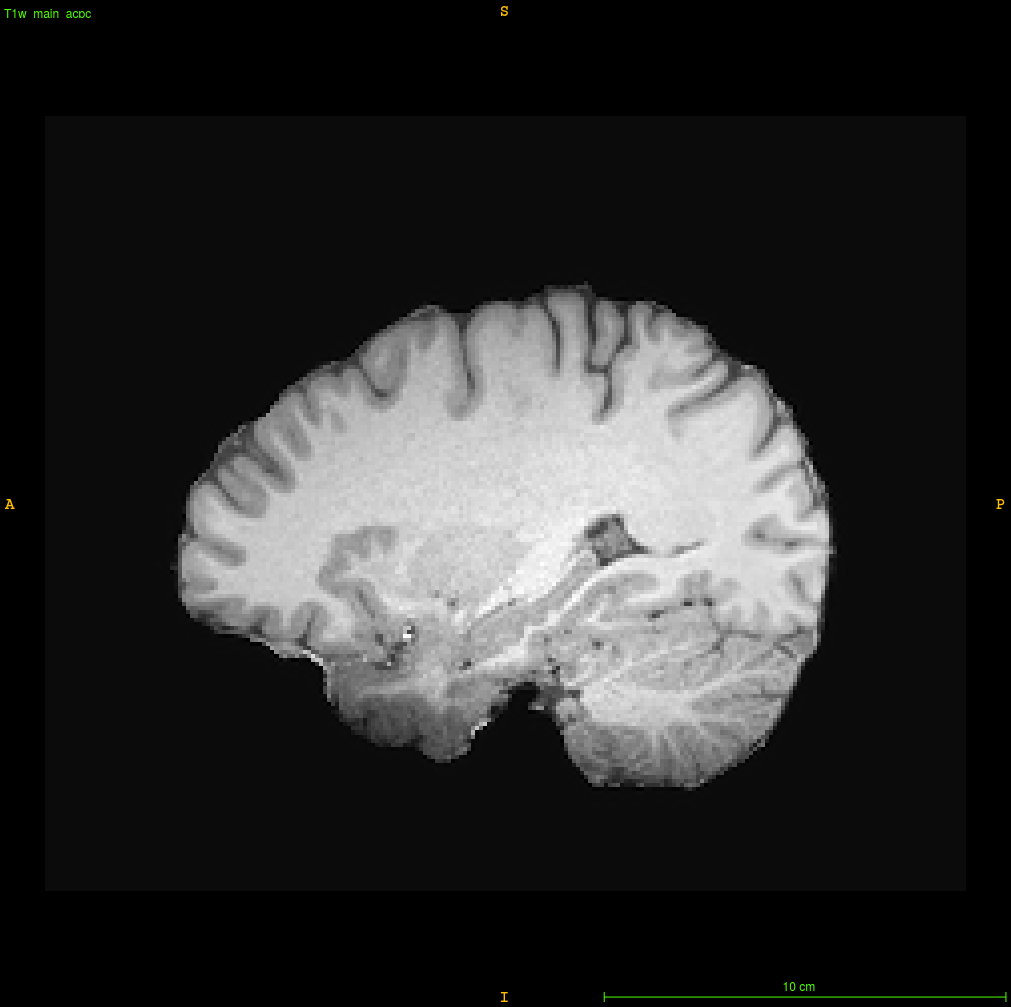
How do you approach correcting greater-than-typical-3T tissue intensity inhomogeneity in structural MRI data? I’ve attached a screenshot illustrating an egregious example that has resisted all of the tools that I typically use (ANTs’s antsAtroposN4.sh, FSL’s fast, various parameters applied for each, use of priors, etc.). The temporopolar regions refuse to normalize their intensity despite my best efforts, and so I would appreciate any suggestions from the community — thanks in advance!
Interesting - have you tried applying N4 twice (once to the output of the previous run)?
1 Like
Worth a shot — I’ll report back.
1 Like
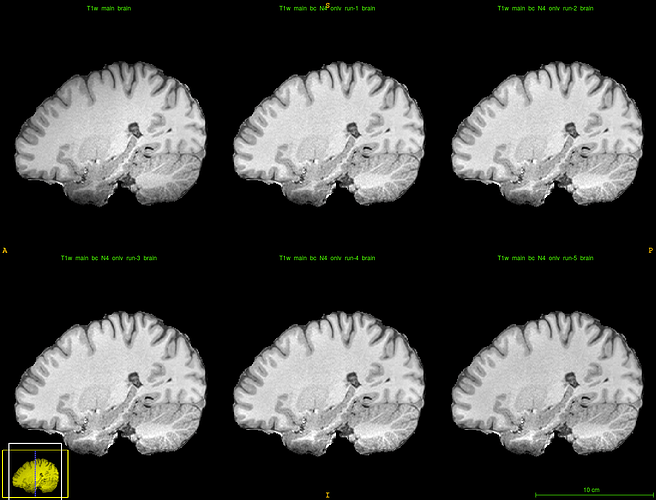
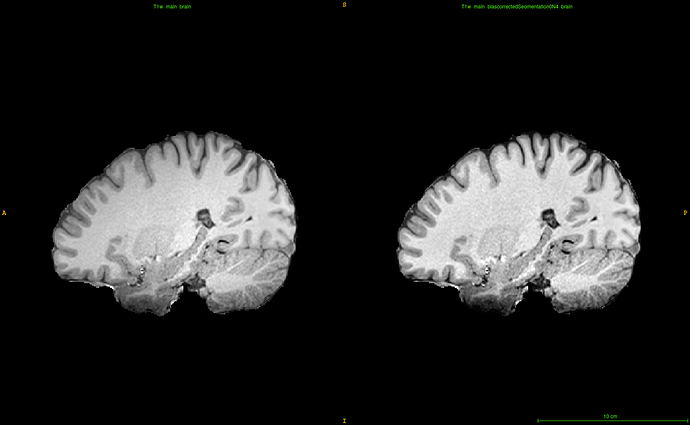
Following @ChrisGorgolewski’s suggestion, I repeatedly applied N4 to same T1 data. The effects were interesting and helpful — feeding the last of these volumes into FreeSurfer produced surfaces that were at least broadly consistent with the contours of the temporal lobe.
So repeated application of ANTs’s N4 bias correction seems to be a solution, while my initial, usual approach of applying antsAtroposN4.sh appeared to exacerbate the problem (below).
Thanks!
1 Like