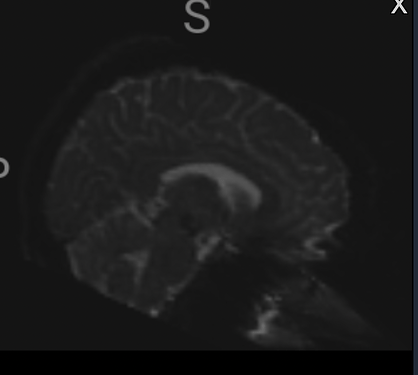
Summary of what happened:
I have a set of diffusion images (max b-value 4900, GRE fmaps) I am preprocessing using QSIprep v.0.16.1. In my pipeline, I preprocess using the input code below and then reconstruct using DSI Studio’s QSDR. After preprocessing, I noticed an artifact in my images on the outside of the brain. I’m concerned about any potential impacts on my analyses. It does appear that the artifact area of the image gets masked out when reconstructing but I would like another opinion before moving forward. Also, I’m wondering if anyone knows why this artifact appears and if it is common with diffusion processing or something in QSIs processing in general.
There are no errors reported during preprocessing.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv \
-B ~/cossio/ \
-B ~/cossio/SNAG/sourcedata:/work \
~ /cossio/Scripts/QSI/sing_containers/qsiprep-0.16.1.sif \
/work \
~/cossio/SNAG/QSI \
participant \
-w ~/cossio/SNAG/QSI/workflow/$id \
--participant_label $sid \
--skip_bids_validation \
--output_resolution 1.8 \
--fs-license-file ~/cossio/Scripts/QSI/license.txt \
--denoise-method dwidenoise \
--dwi-denoise-window 5 \
--hmc_model 3dSHORE \
--hmc-transform Rigid \
--shoreline_iters 2
Version:
qsiprep-0.16.1.sif
Environment (Docker, Singularity / Apptainer, custom installation):
singularity/apptainer built using docker tag qsiprep-0.16.1.sif
Data formatted according to a validatable standard? Please provide the output of the validator:
Relevant log outputs (up to 20 lines):