Hi,
I’m looking for some suggestions on how to go about troubleshooting these artifacts that I see in the fmriprep-report html files from fmriprep 20.2.1. These examples are all from different subjects - but happen in multiple runs somewhat randomly:
A) (brighter vs darker patterns)
B) (intensity difference?)
C)
D) (brighter cortical vs darker subcortical)
- any tips on what could cause these behaviors?
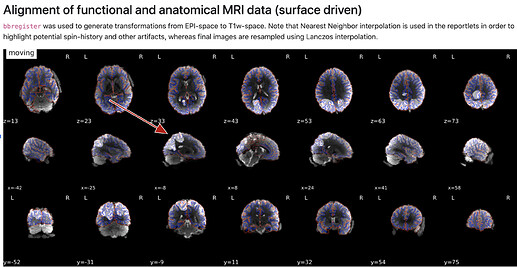
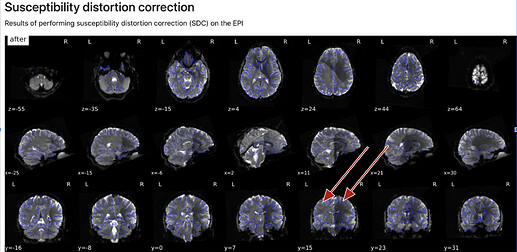
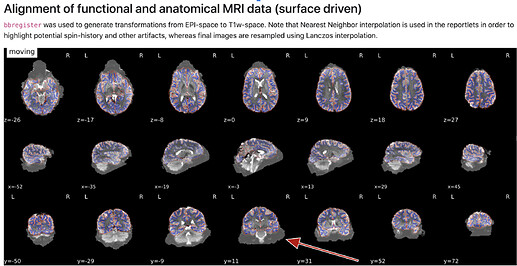
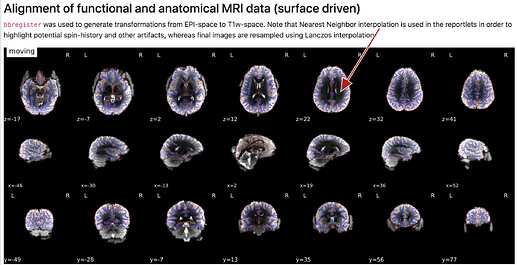
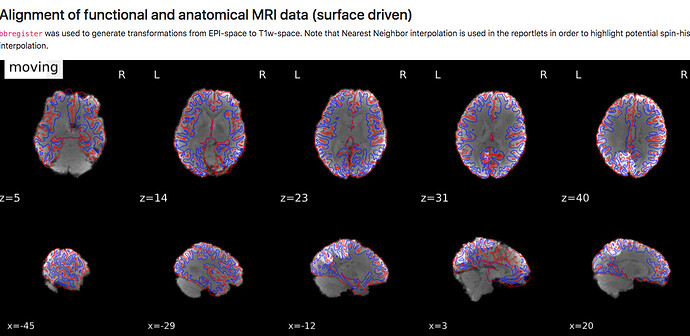
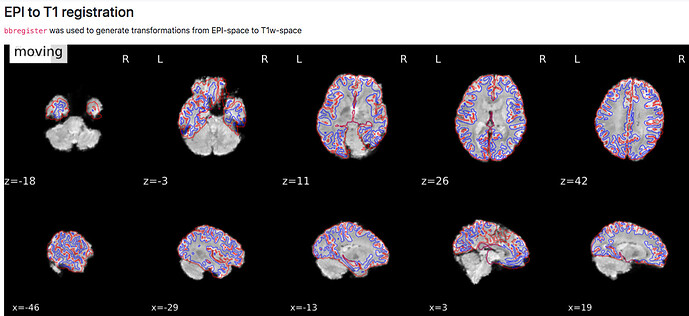
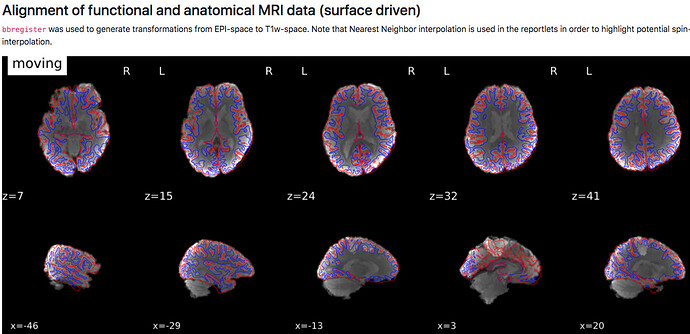
- how are the images used as background calculated, eg for the “alignment of functional and anatomical MRI data (surface driven)” and for the “SDC” plots?
Thanks in advance!
Francesca
forgot to mention: both the raw input to fMRIprep and the output look good from a manual inspection
I have the same question! I have issue A (brightness “ring” around the brain, and some large bright vs. dark patterns) in fMRIprep 20.2.0, but add some additional context.
I have the same subjects run through fMRIprep 1.2.6-1 and fMRIprep 20.2.0 and the older version of fMRIprep does not show issues that look like this in the reports. One additional change with the newer version is the additional of a pediatric template as an output space, but I didn’t expect this change to result from that. Both have pre-run (the same) Freesurfer input. No susceptibility distortion correction applied in either case.
Is this something about the Nearest Neighbor interpolation vs. Lanczos interpolation? Did that change between versions? Or how EPIs are displayed in the report? Is this even something to be concerned about?
Command for 20.2.0:
/usr/local/miniconda/bin/fmriprep /cbica/projects/cbpd_main_data/CBPD_bids /cbica/projects/cbpd_main_data/CBPD_bids/derivatives/fmriprep_t1 participant --participant-label CBPD0172 --fs-license-file /cbica/projects/cbpd_main_data/license.txt --fs-subjects-dir /cbica/projects/cbpd_main_data/CBPD_bids/derivatives/freesurfer_t1 --bids-filter-file /cbica/projects/cbpd_main_data/CBPD_bids/derivatives/fmriprep/ses_01_CBPD0172.json --skull-strip-template MNIPediatricAsym:res-1:cohort-2 --output-spaces MNI152NLin6Asym T1w MNIPediatricAsym:res-1:cohort-2 --ignore sbref --verbose --nthreads 8 -w /tmp
Command for 1.2.6-1
/usr/local/miniconda/bin/fmriprep /cbica/projects/cbpd_main_data/CBPD_bids /cbica/projects/cbpd_main_data/CBPD_bids/derivatives/ participant -w /cbica/projects/cbpd_main_data/fmriprep_wd/ --participant-label sub-CBPD0172 --fs-license-file /home/utooley/license.txt --output-space T1w template --ignore fieldmaps sbref --nthreads 1
20.2.0
1.2.6-1
1 Like
Update: after looking through the final outputs, I’ve decided that the bright-dark spots aren’t directly affecting the BOLD output…but I’m still not sure why it changed. So I’d still be curious about this, because many of my subjects now have a bright “ring” around the brain (see below) in 20.2.0 that I didn’t see in the reports from 1.2.6-1.
1 Like
Hi! I have a similar issue with fmriprep v20.2.1 (parts of the brain are much brighter than the rest due to failure of intensity normalization I suppose?). I am just curious that based on what you decided that the bright-dark spots aren’t directly affecting the BOLD output? Thanks!
Looks like some issues with creating the bold reference file. Do your boldref.nii.gz files show these artifacts as well?
1 Like
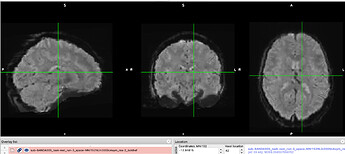
@effigies thanks for looking into this and for the tip! In my case, the four boldref.nii.gz files corresponding to my original screenshots for A, B, C, D look reasonable overall, and don’t show the bright/light pattern I see in the html reports.
This is the one corresponding to A.
That’s a good sign, then, since those should be what’s being plotted. So it’s probably a bug in the plotting itself. If you have a dataset that you can make public that reliably produces these issues, we can try to debug it.
Confirming that I also do not see this issue in the boldref.nii.gz, which is reassuring (I thought it might be something about the plotting, since runs of the exact same data through a previous fmriprep version didn’t show it). However, I don’t believe my dataset can be shared publicly yet–I will check.
1 Like
Unfortunately, I can’t share the data. I could share the logs, is there any specific ones to look into? (btw, with fmriprep 20.2.3 I still get similar behaviors in the html reports).