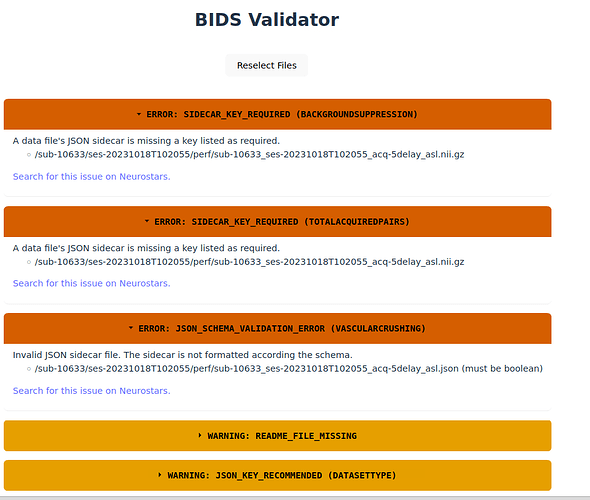
Hi Steven,
Thanks for the quick reply! I have no prior experience with fmriprep or aslprep, so I appreciate any guidance you or others here have for me.
Of course I would have a typo!  But apparently that was not the only issue.
But apparently that was not the only issue.
I did a couple runs:
(1) fixing the indendedFor to IntendedFor, no luck;
(2) additionally removing BIDS URI formatting so that the AP and PA field map json files IntendedFor looks as follows, still no luck.
sub-10709_ses-20240723T113136_dir-AP_epi.json
sub-10709_ses-20240723T113136_dir-PA_epi.json
"IntendedFor": [
"func/sub-10709_ses-20240723T113136_task-rest_bold.nii.gz",
"func/sub-10709_ses-20240723T113136_task-vv2bk_bold.nii.gz",
"func/sub-10709_ses-20240723T113136_task-sceneenc_bold.nii.gz",
"fmap/sub-10709_ses-20240723T113136_acq-5delay_asl.nii.gz"
]
(3) On the third run I removed the --skip_bids_validation flag and added --fmap-bspline, thinking that maybe either the validation or asking it to do bspline would force some sort of an error. Unfortunately that didn’t change anything and it still ran all the way through without doing the distortion correction.
Notably, it did print this:
250313-15:13:46,532 cli INFO:
Making sure the input data is BIDS compliant (warnings can be ignored in most cases).
bids-validator does not appear to be installed
250313-15:14:08,903 nipype.workflow IMPORTANT:
Running ASLPrep version 0.7.5
so it seems a bids-validator is not in the 0.7.5 image.
I searched for “fmap” in the output text and saw this:
250313-13:30:30,89 nipype.workflow IMPORTANT:
ASLPrep started!
250313-13:30:32,203 nipype.workflow INFO:
Workflow aslprep_0_7_wf settings: ['check', 'execution', 'logging', 'monitoring']
250313-13:30:32,701 nipype.workflow INFO:
Running in parallel.
250313-13:30:32,853 nipype.workflow INFO:
[MultiProc] Running 0 tasks, and 19 jobs ready. Free memory (GB): 310.90/310.90, Free processors: 8/8.
250313-13:30:33,137 nipype.workflow INFO:
[Node] Outdated cache found for "aslprep_0_7_wf.sub_10709_wf.bidssrc".
250313-13:30:33,138 nipype.workflow VERBOSE:
[Node] Old/new hashes = ca77c828d8c3c0a04ea4e2ba4a16ff69/4720a8fda2e43a533a583fffd0abae92
250313-13:30:33,156 nipype.workflow VERBOSE:
Some dictionary entries had differing values:
* subject_data: (('asl', (('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/perf/sub-10709_ses-20240723T113136_acq-5delay_asl.nii.gz', '39c68ecababff0afe81ffe9938cb8b96'),)), ('flair', ()), ('fmap', (('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/fmap/sub-10709_ses-20240723T113136_dir-AP_epi.nii.gz', '0d27fb9e93a0eee9490518f544ca5b3f'), ('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/fmap/sub-10709_ses-20240723T113136_dir-PA_epi.nii.gz', '0d006464c0b7619e9995f13a64d0d459'))), ('roi', ()), ('sbref', ()), ('t1w', (('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/anat/sub-10709_ses-20240723T113136_acq-mprage_T1w.nii.gz', '1554f32cfca6552c8532fc698d9a1195'),)), ('t2w', ())) != (('asl', (('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/perf/sub-10709_ses-20240723T113136_acq-5delay_asl.nii.gz', 'f592838b518523fe6e2085414775f9ff'),)), ('flair', ()), ('fmap', (('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/fmap/sub-10709_ses-20240723T113136_dir-AP_epi.nii.gz', '0d27fb9e93a0eee9490518f544ca5b3f'), ('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/fmap/sub-10709_ses-20240723T113136_dir-PA_epi.nii.gz', '0d006464c0b7619e9995f13a64d0d459'))), ('roi', ()), ('sbref', ()), ('t1w', (('/N/scratch/echumin/TEMP-PROJECT/sub-10709/ses-20240723T113136/anat/sub-10709_ses-20240723T113136_acq-mprage_T1w.nii.gz', '39d9360ccbdb2046d868012a6d0c850b'),)), ('t2w', ()))
so it does see the fmap folder and images.
I don’t know is this is useful, but here are all the outputs generated and fmap derivative folder does not exist:
[echumin@i52 ses-20240723T113136]$ ls *
anat:
sub-10709_ses-20240723T113136_acq-mprage_desc-brain_mask.json sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_desc-preproc_dseg.json
sub-10709_ses-20240723T113136_acq-mprage_desc-brain_mask.nii.gz sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_desc-preproc_dseg.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_desc-preproc_T1w.json sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_desc-preproc_T1w.json
sub-10709_ses-20240723T113136_acq-mprage_desc-preproc_T1w.nii.gz sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_desc-preproc_T1w.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_dseg.nii.gz sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_dseg.json
sub-10709_ses-20240723T113136_acq-mprage_from-MNI152NLin2009cAsym_to-T1w_mode-image_xfm.h5 sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_dseg.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5 sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_label-CSF_desc-preproc_probseg.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_label-CSF_probseg.nii.gz sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_label-CSF_probseg.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_label-GM_probseg.nii.gz sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_label-GM_desc-preproc_probseg.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_label-WM_probseg.nii.gz sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_label-GM_probseg.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_desc-brain_mask.json sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_label-WM_desc-preproc_probseg.nii.gz
sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_desc-brain_mask.nii.gz sub-10709_ses-20240723T113136_acq-mprage_space-MNI152NLin2009cAsym_res-2_label-WM_probseg.nii.gz
figures:
sub-10709_ses-20240723T113136_acq-5delay_desc-basilByTissueType_cbf.json sub-10709_ses-20240723T113136_acq-5delay_desc-basilGM_cbf.json sub-10709_ses-20240723T113136_acq-5delay_desc-cbf_cbf.svg
sub-10709_ses-20240723T113136_acq-5delay_desc-basilByTissueType_cbf.svg sub-10709_ses-20240723T113136_acq-5delay_desc-basilGM_cbf.svg sub-10709_ses-20240723T113136_acq-5delay_desc-coreg_asl.svg
sub-10709_ses-20240723T113136_acq-5delay_desc-basil_cbf.json sub-10709_ses-20240723T113136_acq-5delay_desc-carpetplot_asl.svg sub-10709_ses-20240723T113136_acq-5delay_desc-summary_asl.html
sub-10709_ses-20240723T113136_acq-5delay_desc-basil_cbf.svg sub-10709_ses-20240723T113136_acq-5delay_desc-cbfByTissueType_cbf.json sub-10709_ses-20240723T113136_acq-5delay_desc-validation_asl.html
sub-10709_ses-20240723T113136_acq-5delay_desc-basilGMByTissueType_cbf.json sub-10709_ses-20240723T113136_acq-5delay_desc-cbfByTissueType_cbf.svg
sub-10709_ses-20240723T113136_acq-5delay_desc-basilGMByTissueType_cbf.svg sub-10709_ses-20240723T113136_acq-5delay_desc-cbf_cbf.json
perf:
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S1056Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-4S956Parcels_desc-basil_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S1056Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-4S956Parcels_desc-basil_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S1056Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-4S956Parcels_desc-basilGM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S1056Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-4S956Parcels_desc-basilWM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S1056Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Glasser_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S1056Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Glasser_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S156Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Glasser_desc-basil_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S156Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Glasser_desc-basil_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S156Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Glasser_desc-basilGM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S156Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Glasser_desc-basilWM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S156Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Gordon_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S156Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Gordon_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S256Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Gordon_desc-basil_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S256Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Gordon_desc-basil_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S256Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Gordon_desc-basilGM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S256Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Gordon_desc-basilWM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S256Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-HCP_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S256Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-HCP_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S356Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-HCP_desc-basil_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S356Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-HCP_desc-basil_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S356Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-HCP_desc-basilGM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S356Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-HCP_desc-basilWM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S356Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Tian_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S356Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Tian_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S456Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Tian_desc-basil_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S456Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Tian_desc-basil_coverage.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S456Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Tian_desc-basilGM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S456Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_atlas-Tian_desc-basilWM_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S456Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_desc-confounds_timeseries.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S456Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_desc-coreg_aslref.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S556Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_desc-coreg_aslref.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S556Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_desc-hmc_aslref.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S556Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_desc-hmc_aslref.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S556Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_desc-qualitycontrol_cbf.tsv
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S556Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_from-aslref_to-T1w_mode-image_xfm.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S556Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_from-aslref_to-T1w_mode-image_xfm.txt
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S656Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_from-orig_to-aslref_mode-image_xfm.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S656Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_from-orig_to-aslref_mode-image_xfm.txt
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S656Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_aslref.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S656Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_aslref.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S656Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_att.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S656Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_att.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S756Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_cbf.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S756Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_cbf.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S756Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basil_att.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S756Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basil_att.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S756Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basil_cbf.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S756Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basil_cbf.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S856Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basilGM_cbf.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S856Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basilGM_cbf.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S856Parcels_desc-basil_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basilWM_cbf.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S856Parcels_desc-basil_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-basilWM_cbf.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S856Parcels_desc-basilGM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-brain_mask.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S856Parcels_desc-basilWM_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S956Parcels_cbf.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-preproc_asl.json
sub-10709_ses-20240723T113136_acq-5delay_atlas-4S956Parcels_coverage.tsv sub-10709_ses-20240723T113136_acq-5delay_space-MNI152NLin2009cAsym_desc-preproc_asl.nii.gz
I did the last run with -vv and saved the terminal to a text file, in case that may be of use:
aslprep075_run_terminallog-vv.txt (1.2 MB)