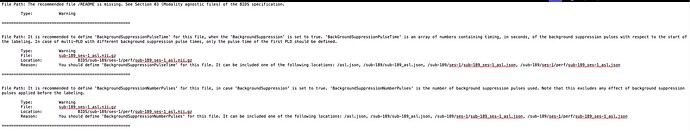
Summary of what happened:
Hello!!!
We are processing pcasl data with aslprep. The process finished successfully, however we found the following error in the log file.
We wanted to check if we are doing properly or if we are failing at any step, and check whether differences are mainly due to basil or aslprep use.
Thank you in advance!
Lucía
Command used (and if a helper script was used, a link to the helper script or the command generated):
Moreover, we wanted to compare outputs from aslprep versus basil itself. We found that though data and parameters introduced are the same results does not seem to be similar. Here I show you argument lines for both:
Aslprep
Input file: /neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/prueba_189/aslprep_wf/single_subject_189_wf/asl_preproc_ses_1_wf/compute_cbf_wf/extract_deltam/sub-189_ses-1_asl_DeltaMOrCBF.nii.gz
Saving results in natve (ASL aquisition) space to /neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/prueba_189/aslprep_wf/single_subject_189_wf/asl_preproc_ses_1_wf/compute_cbf_wf/extract_deltam/native_space
Pre-processing
Number of TIs in list: 9
TIs list: --ti1=2.0 --ti2=2.0 --ti3=2.5 --ti4=2.5 --ti5=3.0 --ti6=3.0 --ti7=3.5 --ti8=3.5 --ti9=3.5
Input ASL format is: diff
Input block format is: rpt
Number of voxels is:442368
Number of repeats in data is:1
Outputting ASL data mean at each TI
Done.
Label-control difference data provided: /neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/prueba_189/aslprep_wf/single_subject_189_wf/asl_preproc_ses_1_wf/compute_cbf_wf/extract_deltam/sub-189_ses-1_asl_DeltaMOrCBF.nii.gz
Number of inversion times: 9
Number of timepoints in data: 9
Number of repeats in data: 1
2.0 2.0 2.5 2.5 3.0 3.0 3.5 3.5 3.5
2.0
Defaulting to brain extracted calibration image as initial registration reference
Using brain extracted calibration image as initial registration reference
Using mask: /neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/prueba_189/aslprep_wf/single_subject_189_wf/asl_preproc_ses_1_wf/compute_cbf_wf/refine_mask/ref_bold_corrected_brain_mask_maths_refinemask.nii.gz
T1: 1.3
T1b: 1.65
Bolus duration: 1.5
Setting up BASIL
cASL model
Infer arterial component
Fixed bolus duration
Variable arterial arrival time
Setting prior/initial (tissue/gray matter) bolus arrival time to 1.3
Setting std dev of the (tissue) BAT prior to 1
Instructing BASIL to use automated spatial smoothing
(Also alpha defined as 0.7)
Basil
oxford_asl -i=/home/jovyan/neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/basilprueba/sub189/ses1/perf/sub-189_ses-1_asl.nii.gz --iaf=tc --ibf=rpt --casl --bolus=1.5 --rpts=1,1,1,1,1,1,1,1,1 --tis=2,2,2.5,2.5,3,3,3.5,3.5,3.5 --fslanat=/home/jovyan/neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/basilprueba/sub189/ses1/anat/T1w.anat -c=/home/jovyan/neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/basilprueba/sub189/ses1/perf/sub-189_ses-1_m0scan.nii.gz --cmethod=single --tr=4.3 --cgain=1 --tissref=csf --t1csf=4.3 --t2csf=750 --t2bl=150 --te=0 -o=/home/jovyan/neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/basilprueba --bat=1.3 --t1=1.3 --t1b=1.65 --alpha=0.7 --spatial=1 --fixbolus --mc --pvcorr --artoff
Version:
aslprep 0.5.0
Environment (Docker, Singularity / Apptainer, custom installation):
Neurodesk
Data formatted according to a validatable standard? Please provide the output of the validator:
Relevant log outputs (up to 20 lines):
stdout 2023-12-04T12:39:59.161239:Generating calibration image in structural space
stderr 2023-12-04T12:39:59.168147:Image Exception : #63 :: No image files match: /tmp/fsl_xu33pve6_ox_asl/struc
stderr 2023-12-04T12:39:59.168454:Error : No image files match: /tmp/fsl_xu33pve6_ox_asl/struc
stderr 2023-12-04T12:39:59.168454:Image Exception : #22 :: Failed to read volume /tmp/fsl_xu33pve6_ox_asl/struc
stderr 2023-12-04T12:39:59.168639:Error : No image files match: /tmp/fsl_xu33pve6_ox_asl/struc
stderr 2023-12-04T12:39:59.168639:Failed to read volume /tmp/fsl_xu33pve6_ox_asl/struc
stdout 2023-12-04T12:39:59.169753:Loading supplied PV images
stdout 2023-12-04T12:39:59.170369:PV GM is: /neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/prueba_189/aslprep_wf/single_subject_189_wf/asl_preproc_ses_1_wf/compute_cbf_wf/gm_tfm/sub-189_ses-1_T1w_corrected_xform_masked_pve_1_trans.nii.gz
stdout 2023-12-04T12:39:59.192364:PV WM is: /neurodesktop-storage/Agueda/BIDS/derivatives/aslprep_Lucia/prueba_189/aslprep_wf/single_subject_189_wf/asl_preproc_ses_1_wf/compute_cbf_wf/wm_tfm/sub-189_ses-1_T1w_corrected_xform_masked_pve_2_trans.nii.gz
stdout 2023-12-04T12:39:59.318393:WARNING: No structural->standard space transformation - cannot generate cortical-only GM/WM masks