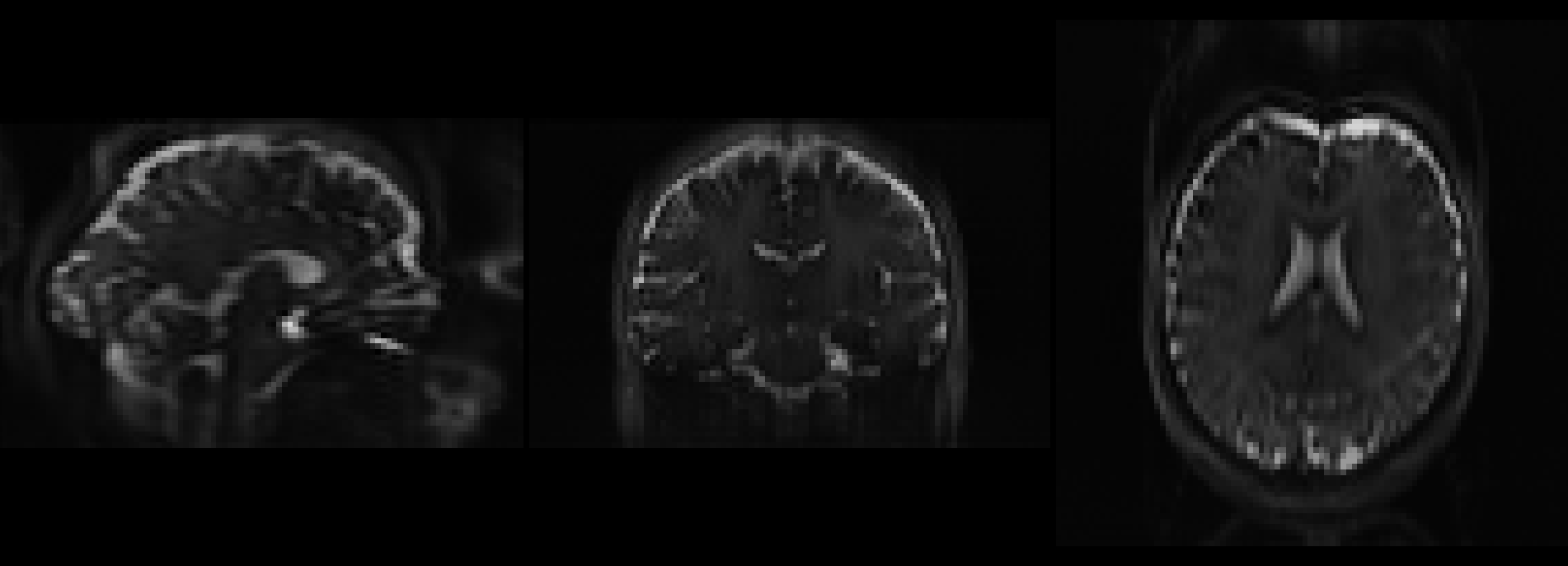
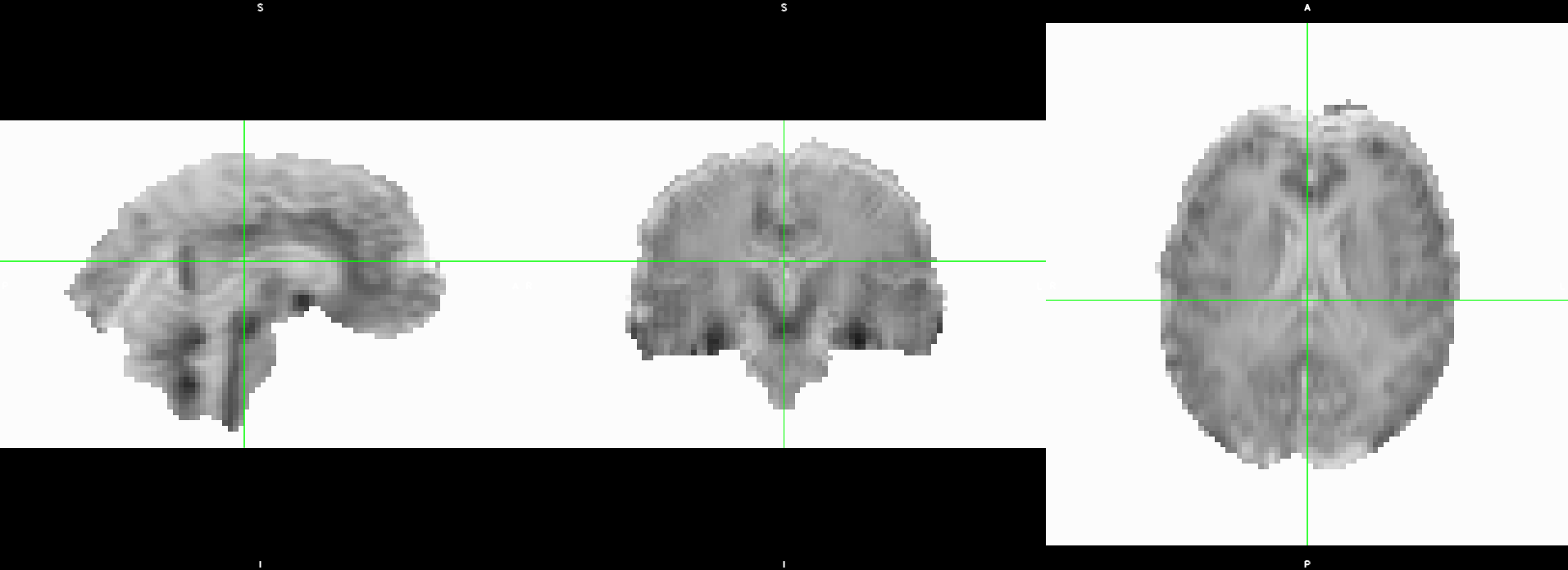
Summary
Hello, the issue I am having is the below pasted error I have encountered for a single subject. I have tried rerunning and adjusting permissions, but only this subject fails, the other 8 ran without errors. Has anyone encountered this keyerror and can provide any info on what’s going on?
Much appreciated,
Caleb
Version:
ASLPrep 0.7.2
Singularity container
Error
Traceback (most recent call last):
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node cbf_summary.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/interfaces/plotting.py", line 164, in _run_interface
).plot()
^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/utils/plotting.py", line 47, in plot
statfile = plot_stat_map(
^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/utils/plotting.py", line 99, in plot_stat_map
svg = extract_svg(display, compress=compress)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/niworkflows/viz/utils.py", line 140, in extract_svg
image_svg = svg2str(display_object, dpi)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/niworkflows/viz/utils.py", line 132, in svg2str
display_object.frame_axes.figure.savefig(
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/figure.py", line 3390, in savefig
self.canvas.print_figure(fname, **kwargs)
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/backend_bases.py", line 2193, in print_figure
result = print_method(
^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/backend_bases.py", line 2043, in
print_method = functools.wraps(meth)(lambda *args, **kwargs: meth(
^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/backends/backend_svg.py", line 1339, in print_svg
self.figure.draw(renderer)
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/artist.py", line 95, in draw_wrapper
result = draw(artist, renderer, *args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/artist.py", line 72, in draw_wrapper
return draw(artist, renderer)
^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/figure.py", line 3143, in draw
artists = self._get_draw_artists(renderer)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/matplotlib/figure.py", line 231, in _get_draw_artists
ax.apply_aspect(locator(ax, renderer) if locator else None)
^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/aslprep/lib/python3.11/site-packages/nilearn/plotting/displays/_slicers.py", line 1572, in _locator
[[left_dict[axes], y0], [left_dict[axes] + width_dict[axes], y1]]
~~~~~~~~~^^^^^^
KeyError: