Summary of what happened:
Hi everyone. I’m attempting to use ASLPrep 25.1.0 to process 2D multi-band PCASL data gathered using the CMRR sequence, but it doesn’t seem to recognize the accompanying spin-echo field maps. I have tried multiple fixes based on other forum posts here, but unfortunately none have worked so far.
I have data from three participants, some with multiple runs, set up in the following way:
sub-XX/ses-XX/perf/sub-XX_ses-XX_dir-PA_run-01_asl.nii.gz
sub-XX/ses-XX/perf/sub-XX_ses-XX_dir-PA_run-01_asl.json
sub-XX/ses-XX/fmap/sub-XX_ses-XX_dir-AP_run-01_epi.nii.gz
sub-XX/ses-XX/fmap/sub-XX_ses-XX_dir-AP_run-01_epi.json
sub-XX/ses-XX/fmap/sub-XX_ses-XX_dir-PA_run-01_epi.nii.gz
sub-XX/ses-XX/fmap/sub-XX_ses-XX_dir-PA_run-01_epi.json
The fmap JSONs contain the following (among other fields):
"PhaseEncodingDirection": "j-",
"TotalReadoutTime": 0.0484496,
"IntendedFor": ["ses-XX/perf/sub-XX_ses-XX_dir-PA_run-01_asl.nii.gz"]
Where j- is used for the AP fmap and j is used for the PA fmap. The perfusion data also includes the field PhaseEncodingDirection: j.
I have tried a few fixes already, including adding or removing the BIDS URI (bids::sub-XX/) in the IntendedFor field, adding or removing brackets around the IntendedFor field, and adding
"B0FieldIdentifier": "fmap1"
"B0FieldSource": "fmap1"
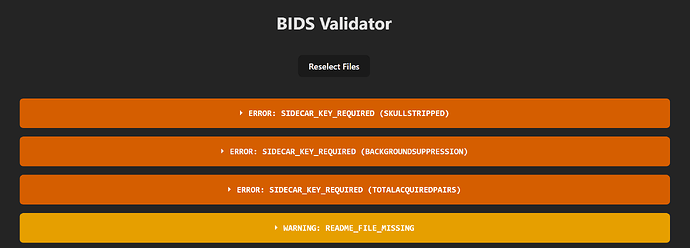
to the fmap and perfusion JSONs, respectively. The online BIDS validator gives errors regarding a few missing keys (BackgroundSuppression, SkullStripped, and TotalAcquiredPairs), but otherwise nothing about field maps. I am running ASLPrep via singularity in Ubuntu via a bash script with the command below:
Command used (and if a helper script was used, a link to the helper script or the command generated):
$(singularity run \
--cleanenv \
--bind ${BIDSINDATA}:/SubjectsMBPCASL_25.1.0 \
--bind ${BIDSOUTDATA}:/SubjectsMBPCASL_25.1.0_derivatives \
--bind /home/Software/FreeSurfer/freesurfer-7.3.2/freesurfer:/fs_license \
--bind /nox2/data/aslprep_workdir_tmp:/workdir \
/home/Software/aslprepS-25.1.0 \
--skip_bids_validation \
/SubjectsMBPCASL_25.1.0 \
/SubjectsMBPCASL_25.1.0_derivatives \
participant \
--participant_label ${SUBJECT} \
-w /workdir/work_sub-${SUBJECT} \
--output-spaces MNI152NLin6Asym:res-2 anat fsaverage \
--omp-nthreads 2 \
--nthreads 4 \
--mem-mb 96000 \
--fs-license-file /fs_license/license.txt \
&> ${LOGDIR}/aslprepS_25.1.0_MBPCASL_Site_${SITE}_${SUBJECT}_${DATEF}.log) &
Version:
ASLPrep 25.1.0
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
See screenshot below.
Relevant log outputs (up to 20 lines):
251209-11:03:29,858 nipype.workflow INFO:
Collected run data for /SubjectsMBPCASL_25.1.0/sub-XX/ses-XX/perf/sub-XX_ses-XX_dir-PA_run-01_asl.nii.gz:
aslcontext: /SubjectsMBPCASL_25.1.0/sub-XX/ses-XX/perf/sub-XX_ses-XX_dir-PA_run-01_aslcontext.tsv
m0scan: null
sbref: null
251209-11:03:29,871 nipype.workflow INFO:
No single-band-reference found for sub-XX_ses-XX_dir-PA_run-01_asl.nii.gz.
251209-11:03:29,956 nipype.workflow INFO:
Stage 1: Adding HMC aslref workflow
251209-11:03:29,961 nipype.workflow INFO:
Stage 2: Adding motion correction workflow
251209-11:03:29,966 nipype.workflow INFO:
No fieldmap correction - skipping Stage 3
Screenshots / relevant information:
BIDS Validator:
The dcm2bids JSON file includes:
{
"id" : "mbPCASL_PA_01",
"datatype": "perf",
"suffix": "asl",
"custom_entities": "dir-PA",
"criteria": {
"SeriesDescription": "mbPCASLhr_PA_01"
},
"sidecar_changes": {
"ArterialSpinLabelingType": "PCASL",
"PostLabelingDelay": 1.64186,
"LabelingDuration": 1.5,
"M0Type": "Included",
"RepetitionTimePreparation": 3.58,
"B0FieldSource": "fmap1"
}
},
{
"datatype": "fmap",
"suffix": "epi",
"custom_entities": "dir-PA",
"criteria": {
"SeriesDescription": "PCASLhr_SpinEchoFieldMap_PA_01"
},
"sidecar_changes" : {
"IntendedFor" : [
"mbPCASL_PA_01"
],
"B0FieldIdentifier": "fmap1"
}
},
{
"datatype": "fmap",
"suffix": "epi",
"custom_entities": "dir-AP",
"criteria": {
"SeriesDescription": "PCASLhr_SpinEchoFieldMap_AP_01"
},
"sidecar_changes" : {
"IntendedFor" : [
"mbPCASL_PA_01"
],
"B0FieldIdentifier": "fmap1"
}
},
Thanks for the help!
Keith G. Jones
Department of Psychiatry and Biobehavioral Sciences
University of California, Los Angeles