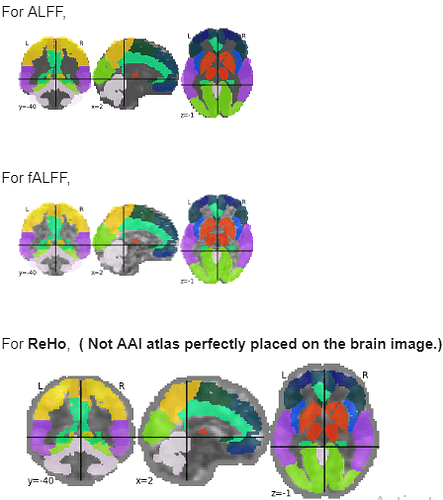
Firstly, I used DPARSF for fMRI preprocessing. It generates ALFF, fALFF, ReHo files for each patient.
Then, I am trying to use plot_roi to plot the aal atlas on the ALFF, fALFF, ReHo files. The aal atlas perfectly places on the ALFF, fALFF files, but it does not work for ReHo files for each patient.
I am using the following codes:
from nilearn import datasets
dataset = datasets.fetch_atlas_aal(version='SPM12')
atlas_filename, labels = dataset.maps, dataset.labels
plotting.plot_roi(atlas_filename, file_name, black_bg= False)
plotting.show()
Can anyone please help me how can I make it perfectly fitted?
Or Can anyone please tell me what are the problems?
Sample fMRI data link: https://drive.google.com/file/d/110uEyTM2nIXC6hdclHk6rJf0tN07-LXV/view?usp=sharing