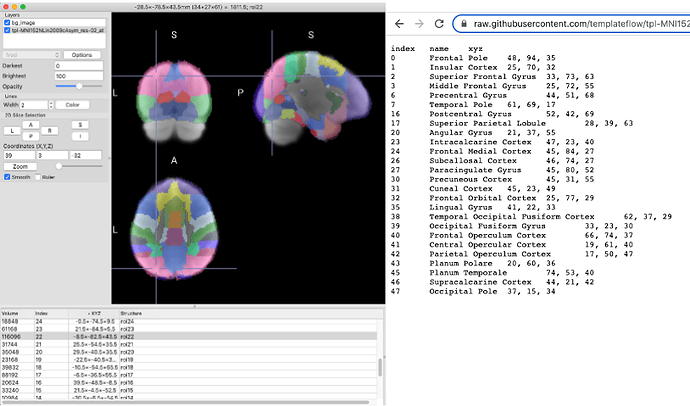
Hi,
These are the commands that I ran:
In [2]: [f.name for f in get('MNI152NLin2009cAsym')]
Out[2]: ['CHANGES',
'LICENSE',
'template_description.json',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-100Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-100Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-200Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-200Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-300Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-300Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-400Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-400Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-500Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-500Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-600Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-600Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-800Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-800Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-1000Parcels7Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-1000Parcels17Networks_dseg.tsv',
'tpl-MNI152NLin2009cAsym_from-MNI152NLin6Asym_mode-image_xfm.h5',
'tpl-MNI152NLin2009cAsym_from-MNIInfant_mode-image_xfm.mat',
'tpl-MNI152NLin2009cAsym_from-OASISTRT20_mode-image_xfm.mat',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPA_desc-th0_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPA_desc-th25_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPA_desc-th50_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPAL_desc-th0_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPAL_desc-th25_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPAL_desc-th50_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOSPA_desc-th0_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOSPA_desc-th25_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-HOSPA_desc-th50_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-100Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-100Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-200Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-200Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-300Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-300Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-400Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-400Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-500Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-500Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-600Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-600Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-800Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-800Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-1000Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-1000Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-brain_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-brain_T1w.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-BrainCerebellumExtraction_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-BrainCerebellumRegistration_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-carpet_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-eye_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-face_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_desc-head_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_label-CSF_probseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_label-GM_probseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_label-WM_probseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_PD.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_T1w.nii.gz',
'tpl-MNI152NLin2009cAsym_res-01_T2w.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPA_desc-th0_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPA_desc-th25_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPA_desc-th50_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPAL_desc-th0_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPAL_desc-th25_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPAL_desc-th50_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_desc-th0_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_desc-th25_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_desc-th50_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_probseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-100Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-100Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-200Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-200Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-300Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-300Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-400Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-400Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-500Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-500Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-600Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-600Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-800Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-800Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-1000Parcels7Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-1000Parcels17Networks_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-brain_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-brain_T1w.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-carpet_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-DKT31_dseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-eye_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-face_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-fMRIPrep_boldref.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_desc-head_mask.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_label-CSF_probseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_label-GM_probseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_label-WM_probseg.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_PD.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_T1w.nii.gz',
'tpl-MNI152NLin2009cAsym_res-02_T2w.nii.gz']
When I go to check the contents of the MNI152NLin2009cAsym directory, this is what I get, some text files with content and a bunch of empty .nii.gz files:
-rw-rw-r-- 1 switt staff 682 22 Jul 19:44 LICENSE
-rw-rw-r-- 1 switt staff 1037 22 Jul 19:44 template_description.json
-rw-rw-r-- 1 switt staff 41703 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-1000Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 38231 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-1000Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 4067 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-100Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 3701 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-100Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 8188 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-200Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 7513 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-200Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 12344 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-300Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 11321 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-300Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 16514 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-400Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 15164 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-400Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 20688 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-500Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 19011 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-500Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 24933 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-600Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 22928 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-600Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 33354 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-800Parcels17Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 30466 22 Jul 19:44 tpl-MNI152NLin2009cAsym_atlas-Schaefer2018_desc-800Parcels7Networks_dseg.tsv
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_from-MNI152NLin6Asym_mode-image_xfm.h5
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_from-MNIInfant_mode-image_xfm.mat
-rw-rw-r-- 1 switt staff 193 22 Jul 19:44 tpl-MNI152NLin2009cAsym_from-OASISTRT20_mode-image_xfm.mat
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_PD.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_T1w.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_T2w.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPAL_desc-th0_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPAL_desc-th25_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPAL_desc-th50_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPA_desc-th0_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPA_desc-th25_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOCPA_desc-th50_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOSPA_desc-th0_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOSPA_desc-th25_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-HOSPA_desc-th50_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-1000Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-1000Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-100Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-100Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-200Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-200Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-300Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-300Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-400Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-400Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-500Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-500Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-600Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-600Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-800Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_atlas-Schaefer2018_desc-800Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-BrainCerebellumExtraction_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-BrainCerebellumRegistration_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-brain_T1w.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-brain_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-carpet_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-eye_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-face_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_desc-head_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_label-CSF_probseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_label-GM_probseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-01_label-WM_probseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_PD.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_T1w.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_T2w.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPAL_desc-th0_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPAL_desc-th25_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPAL_desc-th50_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPA_desc-th0_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPA_desc-th25_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOCPA_desc-th50_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_desc-th0_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_desc-th25_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_desc-th50_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-HOSPA_probseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-1000Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-1000Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-100Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-100Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-200Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-200Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-300Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-300Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-400Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-400Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-500Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-500Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-600Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-600Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-800Parcels17Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_atlas-Schaefer2018_desc-800Parcels7Networks_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-DKT31_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-brain_T1w.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-brain_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-carpet_dseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-eye_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-fMRIPrep_boldref.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-face_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_desc-head_mask.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_label-CSF_probseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_label-GM_probseg.nii.gz
-rw-rw-r-- 1 switt staff 0 22 Jul 19:44 tpl-MNI152NLin2009cAsym_res-02_label-WM_probseg.nii.gz
Please let me know if you need any other details.
Thanks again,
suzanne