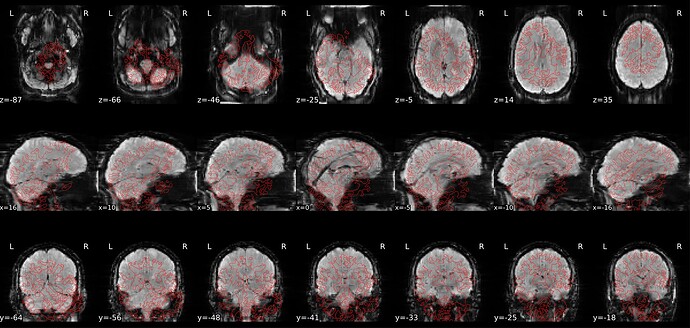
Summary of what happened:
Hi everyone!
We’ve run fMRIPrep using two versions: 23.2.1 and 25.1.1. In the newer version (25.1.1), we’ve noticed that the alignment between the reference EPI (BOLD) image and the anatomical (T1-weighted) image is severely misaligned in just a few cases. As shown in the attached screenshot, the registration is completely off—the image appears noticeably tilted.
Interestingly, these same cases aligned well when processed with fMRIPrep 23.2.1. While we’d prefer to stick with the newer version since it worked well for the majority of our dataset, we’re hoping to find a fix for these misaligned cases.
Has anyone encountered a similar issue with fMRIPrep 25.1.1 or have any suggestions on how to resolve it? Thanks a lot in advance!
Command used (and if a helper script was used, a link to the helper script or the command generated):
# Run fmriprep
apptainer run --cleanenv ${image} ${in} ${out} participant \
--participant_label ${SUBJ} \
--output-spaces MNI152NLin6Asym MNI152NLin2009cAsym T1w fsnative \
--nthreads 12 --mem_mb 64000 \
-w ${prep_work} \
--fs-license-file ./license.txt \
Version:
fMRIPrep 25.1.1 & fMRIPrep 23.2.1
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity/Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
Our dataset is formatted according to the BIDS convention, and everything runs smoothly otherwise.