Hi,
I’m trying to use the nilearn.plotting.plot_surf_stat_map function plot some parcellation using HCPs S1200.L.midthickness_MSMAll.32k_fs_LR.surf.gii as bgmap.
My command is
plotBrainOpts = dict(surf_mesh='S1200.L.pial_MSMAll.32k_fs_LR.surf.gii', hemi='left', view='lateral',
stat_map=somemap, cmap=cmap, bg_on_data=True
bgmap='S1200.L.midthickness_MSMAll.32k_fs_LR.surf.gii', darkness=0.2)
plot_surf_stat_map(**plotBrainOpts)
I’m getting this(partial view):
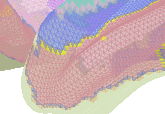
My question is - why do I see the mesh and not its shading on the statmap?
Thx!
is there an alpha channel in the cmap? try removing it. you won’t be able to see the background using transparency, but you can try to use the bg_on_data parameter
I tried - and also tried without the bg_on_data but with alpha=0.5 either as input argument or as cmap value - or both. Always give the same netty structure.
Still thanks…
alpha channel applies to the faces, not the mesh so if the faces become transparent the edges will appear. If you want the brain to appear transparent you can use an alpha channel with view_img_on_surf or view_surf_stat_map . but if what you want is for the sulcal depth to appear set alpha channel to 1 and use bg_on_data
I understood. When it didn’t work, I tried all other permutation out of despair…
However, I think it’s something in the bg file. I tried to use fsaverage.sulc_right of nilearn instead (with fsaverage.pial_right as base and taking some 10242 elements of the stat vector) and it plotted well (of course scattered because the stat in the new space is meaningless).
So next is to understand where the difference (between fsaverage.sulc_right and S1200.L.midthickness_MSMAll.32k_fs_LR.surf.gii) lies…
