Hi Albert. That is strange. I just tried and had no issues. Are you using the version of JHU that is inside BCBtoolkit? Are you using a space in the directory or the name of your files? Can you provide me with the log file?
cheers
mich
Hi Albert. That is strange. I just tried and had no issues. Are you using the version of JHU that is inside BCBtoolkit? Are you using a space in the directory or the name of your files? Can you provide me with the log file?
cheers
mich
When Tractotron calculates the probability and proportion of tract intersection with a lesion does it control for the volume of the tracts? If not, how would you suggest controlling for tract size in calculations using Tractotron outputs? Thank you!
Hi,
I am using the version of JHU that is inside BCBtoolkit. The directory is named “results_x”.
I am trying to upload the logtractrotron but it tells me that since I am a new user I cannot attach files. Do you have an email address where I could send it?
Thank you very much for your help!
Albert
Hi Isaiah,
I don’t think controlling for tract size is the best approach. You should rather control for lesion size. In any case, you can extract an approximate tract size by thresholding the tracts maps in BCBtoolkit at 0.5 and calculating their volumes. Let me know if you manage to do so. cheers. mich
Dear Mich,
Thank you so much for your advice. I agree and do control for lesion size in analyses. At the same time, I wonder if it may also be important to control for tract size. Given the Tractotron is based on probability of intersection, larger tracts have an increased likelihood of intersection with lesions. Let’s say for example a given syndrome is associated with uncinate disconnection, many analyses could also show intersection with ILF and IFOF given they are larger and may give the impression that the syndrome is associated with disconnection of those tracts. Alternatively, if lesions were randomly distributed throughout the brain and you ran an analysis of that hypothetical syndrome you may reach the conclusion that intersection with larger tracts was associated with the syndrome given they are larger and more likely to intersect with lesions but the location might not in essence matter.
Thank you so much for your thoughts,
Isaiah
Sorry, Isaiah, your message was during my holiday, and I forgot to reply when I came back.
This is a good question. However bigger tracts are also more likely to be hit by lesions in patients that do not show your deficit of interest so I would still think that tract size is not a nuisance covariate.
cheers
mich
Hello,
I am using BCBtoolkit for disconnectome maps for the first time and I am running into an error. I’m upload a 2D binary lesion into the “lesion folder” and when I run the map I get the following message:
Can you help?
Thanks
Best,
Nelson
Hi Nelson,
It looks like there is an issue with your lesion file.
Is it in the MNI152? can you share this file with us.
More information on how to use BCBtoolkit can be found in the BCBtoolkit manual available at www.bcblab.com
cheers
mich
Hi,
I have two questions regarding the tractotron tool.
Thanks so much in advance!
Good question that resource is not available anymore online (this is not from our lab…) so I wrote it manually below for you. Note that I noticed that tract do not systematically have their equivalent in teh other hemisphere.
CC_CingG.nii.gz - Corpus callosum connecting the cingulate gyri
CC_Cu.nii.gz - Corpus callosum connecting the cuneus
CC_LG.nii.gz - Corpus callosum connecting the lingual gyri
CC_MFG.nii.gz - Corpus callosum connecting the middle frontal gyri
CC_MOG.nii.gz - Corpus callosum connecting the middle occipital gyri
CC_PoCG.nii.gz - Corpus callosum connecting the postcentral gyri
CC_PrCG.nii.gz - Corpus callosum connecting the precentral gyri
CC_PrCu.nii.gz - Corpus callosum connecting the precuneus gyri
CC_RG.nii.gz - Corpus callosum connecting the rectus gyri
CC_SFG.nii.gz - Corpus callosum connecting the superior frontal gyri
CC_SOG.nii.gz - Corpus callosum connecting the superior occipital gyri
CC_SPG.nii.gz - Corpus callosum connecting the superior parietal lobule
CGC_L.nii.gz - Cingulum Dorsal branch left
CGC_R.nii.gz - Cingulum Dorsal branch right
CGH_L.nii.gz - Cingulum ventral branch left
CGH_R.nii.gz - Cingulum ventral branch right
CST_L.nii.gz - Cortico-Spinal Tract left
CST_R.nii.gz - Cortico-Spinal Tract right
IFO_L.nii.gz - Inferior fronto occipital fasciculus left
IFO_R.nii.gz - Inferior fronto occipital fasciculus right
ILF_L.nii.gz - Inferior longitudinal fasciculus left
ILF_R.nii.gz - Inferior longitudinal fasciculus right
SAF_AG_MOG_L.nii.gz - Small associative fibres angular - middle occipital gyri left
SAF_AG_MOG_R.nii.gz - Small associative fibres angular - middle occipital gyri right
SAF_AG_SMG_L.nii.gz - Small associative fibres angular - supramarginal left
SAF_AG_SMG_R.nii.gz - Small associative fibres angular - supramarginal right
SAF_CingG_PrCG_L.nii.gz - Small associative fibres cingulum - precentral gyri left
SAF_CingG_PrCG_R.nii.gz - Small associative fibres cingulum - precentral gyri right
SAF_CingG_PrCu_L.nii.gz - Small associative fibres cingulum - precuneus left
SAF_CingG_PrCu_R.nii.gz - Small associative fibres cingulum - precuneus right
SAF_CingG_SFG_L.nii.gz - Small associative fibres cingulum - superior frontal gyri left
SAF_CingG_SFG_R.nii.gz - Small associative fibres cingulum - superior frontal gyri right
SAF_Cu_LG_L.nii.gz - Small associative fibres cuneus - lingual gyri left
SAF_Cu_LG_R.nii.gz - Small associative fibres cuneus - lingual gyri right
SAF_Cu_MOG_L.nii.gz - Small associative fibres cuneus - middle occipital gyri left
SAF_Cu_MOG_R.nii.gz - Small associative fibres cuneus - middle occipital gyri right
SAF_Cu_SOG_L.nii.gz - Small associative fibres cuneus - sup. occipital gyri left
SAF_Cu_SOG_R.nii.gz - Small associative fibres cuneus - sup. occipital gyri right
SAF_Fu_IOG_L.nii.gz - Small associative fibres fusiform - superior occipital gyri left
SAF_Fu_IOG_L.nii.gz - Small associative fibres fusiform - superior occipital gyri right
SAF_Fu_MOG_L.nii.gz - Small associative fibres fusiform - inferior occipital gyri left
SAF_IFG_PrCG_L.nii.gz - Small associative fibres inferior frontal - precentral gyri left
SAF_IFG_PrCG_R.nii.gz - Small associative fibres inferior frontal - precentral gyri right
SAF_IOG_MOG_L.nii.gz - Small associative fibres inferior occipital - middle occipital gyri left
SAF_IOG_MOG_R.nii.gz - Small associative fibres occipital - middle occipital gyri right
SAF_ITG_MTG_L.nii.gz - Small associative fibres inferior temporal gyrus - middle temporal gyri left
SAF_ITG_MTG_R.nii.gz - Small associative fibres inferior temporal gyrus - middle temporal gyri right
SAF_LFOG_MFOG_L.nii.gz - Small associative fibres no idea what this is SAF_MFG_IFG_L.nii.gz - Small associative fibres middle frontal - inferior frontal gyri left
SAF_MFG_PrCG_L.nii.gz - Small associative fibres middle frontal - precentral gyri left
SAF_MFG_PrCG_R.nii.gz - Small associative fibres middle frontal - peecental gyri right
SAF_PoCG_SMG_L.nii.gz - Small associative fibres post-central - supramarginal gyri left
SAF_PoCG_SMG_R.nii.gz - Small associative fibres post-central - supramarginal gyri right
SAF_PrCG_PoCG_L.nii.gz - Small associative fibres precentral - postcentral gyri left
SAF_PrCG_PoCG_R.nii.gz - Small associative fibres precentral - postcentral gyri right
SAF_SFG_IFG_L.nii.gz - Small associative fibres superior frontal - inferior frontal l gyri left
SAF_SFG_MFG_L.nii.gz - Small associative fibres superior frontal - middle frontal gyri left
SAF_SFG_MFG_R.nii.gz - Small associative fibres superior frontal - middle frontal gyri right
SAF_SFG_PrCG_L.nii.gz - Small associative fibres superior frontal - precentral gyri left
SAF_SFG_PrCG_R.nii.gz - Small associative fibres superior frontal - precentral gyri right
SAF_SFG_RG_R.nii.gz - Small associative fibres superior frontal - rectus gyri left
SAF_SOG_MOG_L.nii.gz - Small associative fibres superior occipital - middle occipital gyri left
SAF_SOG_MOG_R.nii.gz - Small associative fibres superior occipital - middle occipital gyri right
SAF_SPG_AG_L.nii.gz - Small associative fibres angular - superior parietal gyri left
SAF_SPG_AG_R.nii.gz - Small associative fibres angular - superior parietal gyri right
SAF_SPG_MOG_L.nii.gz - Small associative fibres angular - middle occipital gyrus left
SAF_SPG_PoCG_L.nii.gz - Small associative fibres Superior parietal lobule - postcentral gyrus left
SAF_SPG_PoCG_R.nii.gz - Small associative fibres Superior parietal lobule - postcentral gyrus right
SAF_SPG_PrCu_L.nii.gz - Small associative fibres Superior parietal lobule - precuneus gyrus left
SAF_SPG_PrCu_R.nii.gz - Small associative fibres Superior parietal lobule - precuneus gyrus right
SAF_SPG_SMG_L.nii.gz - Small associative fibres Superior parietal lobule - supramarginal gyrus left
SAF_SPG_SMG_R.nii.gz - Small associative fibres Superior parietal lobule - supramarginal gyrus right
SAF_SPG_SOG_L.nii.gz - Small associative fibres Superior parietal lobule - superior occipital gyrus left
SAF_STG_MTG_L.nii.gz - Small associative fibres Superior parietal lobule - middle temporal gyrus left
SAF_STG_MTG_R.nii.gz - Small associative fibres Superior parietal lobule - middle temporal gyrus left
SAF_STG_SMG_L.nii.gz - Small associative fibres Superior temporal - supramarginal gyri left
SAF_STG_SMG_R.nii.gz - Small associative fibres Superior temporal - supramarginal gyri right
SLF_FP_L.nii.gz - superior longitudinal fasiculus fronto-parietal branch left
SLF_FP_R.nii.gz - superior longitudinal fasiculus fronto-parietal branch right
SLF_FT_L.nii.gz - superior longitudinal fasiculus fronto-temporal branch left
SLF_FT_R.nii.gz - superior longitudinal fasiculus fronto-temporal branch right
SLF_PT_L.nii.gz - superior longitudinal fasiculus parieto-temporal branch left
SLF_PT_R.nii.gz - superior longitudinal fasiculus parieto-temporal branch right
TH_MFG_L.nii.gz - Thalamic projections to middle frontal gyrus left
TH_MFG_R.nii.gz - Thalamic projections to middle frontal gyrus right
TH_MOG_L.nii.gz - Thalamic projections to middle occipital gyrus left
TH_MOG_R.nii.gz - Thalamic projections to middle occipital gyrus right
TH_PoCG_L.nii.gz - Thalamic projections to postcentral gyrus left
TH_PoCG_R.nii.gz - Thalamic projections to postcentral gyrus right
TH_PrCG_L.nii.gz - Thalamic projections to precentral gyrus left
TH_PrCG_R.nii.gz - Thalamic projections to precentral gyrus right
TH_PrCu_L.nii.gz - Thalamic projections to precuneus gyrus left
TH_PrCu_R.nii.gz - Thalamic projections to precuneus gyrus right
TH_SFG_L.nii.gz - Thalamic projections to superior frontal gyrus left
TH_SFG_R.nii.gz - Thalamic projections to superior frontal gyrus right
TH_SOG_L.nii.gz - Thalamic projections to superior occipital gyrus left
TH_SOG_R.nii.gz - Thalamic projections to superior occipital gyrus right
TH_SPG_L.nii.gz - Thalamic projections to superior parietal lobule left
TH_SPG_R.nii.gz - Thalamic projections to superior parietal lobule right
UNC_L.nii.gz - Uncinate left
UNC_R.nii.gz - Uncinate right
As for your additional map, this might be a format issue. Plus if it is 0 or 1 you will definitely have only 1 or 0 in your excel spreadsheet for probability and a continuous value in the proportion.
Maybe you can fix the format using the following code.
for a in *.nii.gz
do echo $a
echo “I love BCBtoolkit”
/Applications/BCBToolKitOSX/Tools/binaries/bin/flirt -interp nearestneighbour -in $a -ref /Applications/BCBToolKitOSX/Tools/extraFiles/MNI152.nii.gz -out r00$a.nii -usesqform -applyxfm
/Applications/BCBToolKitOSX/Tools/binaries/bin/fslmaths r00$a -mul r00$a -bin r00$a
done
let me know if it works.
cheers
mich
Hi again,
thanks for your fast response. I had already coregistered the tract masks using SPM, so afterall, all it took was removing nan values using the fslmaths -nan flag. Now, tractotron gives binary probability values and continuous proportion values as you suggested.
Best, Ida
Hi,
I’m trying to launch the BCBToolkit using Ubuntu, which I installed via WSL2 on Windows.
But when I run BCBToolKit.sh, I get the following error:
Hi jwkw,
It seems to be an issue with Java. I never tested the BCBToolkit with the WSL2 on windows and I am not sure how it interacts with Windows. The issue might be that the compiled binaries (and java resources) are compiled for Linux. If WSL2 uses resources compiled for Windows I don’t think it can work.
Do you have the possibility to install a regular virtual machine? That might be easier to use the BCBToolkit if you only have access to a windows computer
Hope that helps,
Chris
Hi,
I am a beginner in MRI analysis and I would like to know the procedure for the pre-processing of disconnectome mapping.
Which software is used and which pre-processing is necessary before the data can be entered into BCBToolKit’s Disconnectome analysis?
If possible, a simple process would be helpful.
Hi,
I have been trying to use Disconnectome Map in BCB Toolkit on my macOS Ventura 13, but it doesn’t seem to work, the task finishes in less than a second and only produces an empty logs file in the output directory.
The tractotron works very well with the same Nifti lesion mask, i don’t have a clue why it won’t work.
I have reinstalled BCB Tollkit, FSL etc …
Have you any idea ?
Thank you in advance,
Benjamin
Hi Benjamin,
Thanks for your interest.
It looks like you don’t have any tract population in your folder or no nifti in the input of disconnectome. Are you using binary lesions 1x1x1 mm mni152?
Did you download your bcbtoolkit on www.bcblab.com?
Did you try to run the code “sudo master disable” as specified at the beginning of the manual?
Let me know so that I can understand where the problem comes from.
Cheers
Mich
Hi Sawai,
Thanks for your message. Your question regards fundamental information that you will find in Foulon et al. Gigascience 2008.
Good luck
Mich
Hi mich,
Thanks for your answer
I am using a binary lesion file defined in the MNI, that seems appropriate since it can be run in tractotron. Here is a screenshot of the command fslinfo on my lesion mask
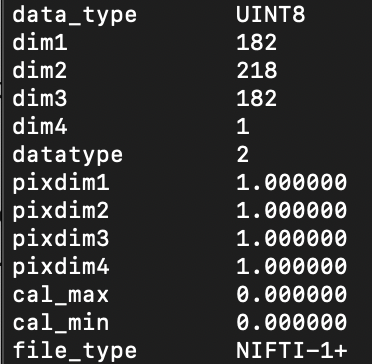
Yes i downloaded BCBToolkit from bcb.lab.com and reinstalled it yesterday.
Tracts are in the “tracts” folder as nii.gz and in tools/extraFiles/tracks as .trk files all in the BCBToolkit folder.
I just ran the comand “sudo spctl --master-disable” as in the manual but disconnectome map doesn’t work better.
Best, Benjamin
That is very strange, I tried it on Ventura, and it usually works. Do you have a space in your directory somewhere or in the name of the files?
If not, I can offer you to run the disconnectome for you here or schedule a Zoom to look at this closer.
Cheers
mich
Thanks for your help,
Can i send you 1 binarized lesion mask as an example so you can try disconnectome yourself and try to tell me where it went wrong ? I tried with several files on several computers with Linux but have the same error.
Once again, Tractotron works well with those files.
Best,
Benjamin
MANAGED BY INCF