OK I just updated the website you should be able to a access all the files by clicking on open data on the first page of www.bcblab.com let me know how it goes :). mich
Yes,I can access the files and download them.Thank you very much,Michel!
Good evening Chris and Michel,
I am starting to use BCB toolkit and I have a question: is it possible to automatically subtract the voxels of the lesion (thus remove the volume of the lesion) from the disconnection map that the software generates?
Thank you in advance,
Yours sincerely
Giulio
Hi Giulio, Thanks for your question. Yes you can with fslmaths (originally from fsl but if you don’t have it you can find a copy in BCBtoolkit.
fslmaths yourlesion -add 1 -uthr 1 tmp; fslmaths yourdisconnectome -mas tmp yourdisconnectomewithoutlesion
Let me know if it works.
mich
Good evening Chris and Michel,
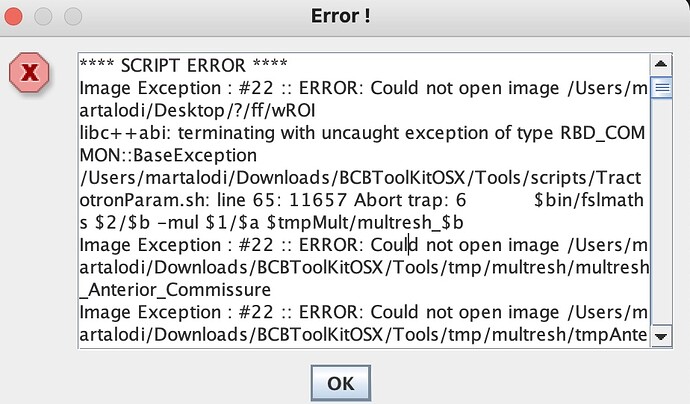
I am starting to use BCB toolkit and I have some problems with Tractotron.
I’m using a nii.gz normalize MNI 152 lesion, but the analysis not run and an icon shows script error (see image)
How I can resolve this problem? I try to use normalisation, but it does not work for a similar reason.
Thank you for the help
Apologies for the delay. I am on a business trip.
Most likely there is a space in the path to BCBtoolkit or the file you would like to process.
Would you have the kindness to share the log file with us please (created in the same folder as the file you would like to process)?
Thanks
mich
Hi Chris and Michel,
I have multiple questions on the structural disconnection analysis using BCB toolkit.
I would like to do a group-level analysis between stroke patients with the presence vs. absence of a certain symptom using tractotron and ANACOM2 as described in Foulon et al. (2018).
-
I ran tractotron and used the output probability data to do statistical testing. I categorized my data as “disconnected” and “spared” depending on the presence or absence of the symptom and used disconnectivity per tract as a continuous variable. Reading your exemplary analysis of category fluency using BCB toolkit (Foulon et al., 2018), how did you categorize the patients with a continous behavioral performance in “disconnected” and “spared” in your Kruskal-Wallis tests using the tractotron output? (Foulon et al., 2018, p. 4 Figure 1), was everything above 0 considered disconnected?
-
Regarding AnaCOM2:
How long do computations last for approximately 20 lesion masks? (working on a MAC with 32GB) Can one speed up the tool by inputting disconnectome maps from the Disconnectome Maps toolkit (in case one wants to run multiple AnaCOM2 settings on the same data)?
I do not have control data. To clarify: If I enter a “published normative value” of 10 (because that means in my context symptom-free), and choose post hoc tests of Disconnected vs. Spared, are then patients with score = 10 (in patient data csv file) considered as “spared”?
- In Foulon et al. (2018), a normative connectome was used on the basis of 10 healthy subjects. In Thiebaut de Schotten, Foulon, and Nachev (2020) and Dulyan et al. (2022), you used the DWI dataset of the Human Connectome Project (Vu et al. 2015). I am curious, how can I change the normative connectome used in BCB toolkit’s Disconnectome Maps?
Thank you so much for taking the time!
Best,
Ida
Hi Ida,
Thanks for your interest.
1/ We chose 50% as a treshold between disconnected and spared.
2/ AnaCom2 is rather long to process data you might want to leave it work overnight . Don’t forget to binarise your disconnectomes.
“published normative value” of 10 means that your statsitical outcome will show you clusters that are significantly different from 10 (10 being the healthy value)
3/ You can change the disconnectome by downloading data on www.bcblab.com (open data) either
DISCONNECTOME Package 1 (+ 20 subjects cardiac gated from Paris) – Foulon et al. Gigascience 2018
DISCONNECTOME Package 2 (15 additional subjects cardiac gated from Paris) – Foulon et al. Gigascience 2018
DISCONNECTOME Package X (180 additional subjects HCP7T 1mm) – Savalaggio et al. Brain 2021
DISCONNECTOME Package X (180 additional subjects HCP7T 2mm) – Thiebaut de Schotten et al. Nat Comm 2020
All you have to do is replace the .trk in BCBToolKitOSX/Tools/extraFiles/tracks with the one downloaded on our website.
Hope this helps.
Kind regards
mich
Thanks, very helpful!
One thing, what do you mean by “do not forget to binarise your disconnectomes”? In the AnaCOM2 tool, I input my lesion directory with binarized lesion masks (MNI space,1mm) as stated in the BCBtoolkit_doc.pdf, right? Do you mean it is possible to input disconnectome maps in the lesion directory, as long as they are binarized? What are the benefits of either approach?
Best, Ida
Hi Ida,
Putting your lesion in AnaCOM2 will provide you with the significant lesioned areas associated with your outcome. If you put your disconnectome, it will calculate for you the disconnected areas associated with your outcome. I just checked with Chris, and you don’t need to binarise data AnaCOM2 will do it for you. Data are binarised because the test will contrast, for each cluster, the effect of direct damage or not (for lesions) or disconnections or not (for disconnectome). Make sure that when you run your disconnectome, they are thresholded at 50% (this is in the interface).
Hi all!
I was using the Tractotron analysis with my dataset of 60 subjects with 1mm MNI-normalized lesions and it keeps throwing me an error of inconsistent orientations. I am using the JHU atlas for the analysis.
I also tried with 1 only subject, and it also gives me the same error.
The tractotron analysis work finely if I put the Tracts directory instead of JHU.
Could you please help me with this and suggest what changes should I make?
Thank you in advance!
Albert
Hi Albert. That is strange. I just tried and had no issues. Are you using the version of JHU that is inside BCBtoolkit? Are you using a space in the directory or the name of your files? Can you provide me with the log file?
cheers
mich
When Tractotron calculates the probability and proportion of tract intersection with a lesion does it control for the volume of the tracts? If not, how would you suggest controlling for tract size in calculations using Tractotron outputs? Thank you!
Hi,
I am using the version of JHU that is inside BCBtoolkit. The directory is named “results_x”.
I am trying to upload the logtractrotron but it tells me that since I am a new user I cannot attach files. Do you have an email address where I could send it?
Thank you very much for your help!
Albert
Hi Isaiah,
I don’t think controlling for tract size is the best approach. You should rather control for lesion size. In any case, you can extract an approximate tract size by thresholding the tracts maps in BCBtoolkit at 0.5 and calculating their volumes. Let me know if you manage to do so. cheers. mich
Dear Mich,
Thank you so much for your advice. I agree and do control for lesion size in analyses. At the same time, I wonder if it may also be important to control for tract size. Given the Tractotron is based on probability of intersection, larger tracts have an increased likelihood of intersection with lesions. Let’s say for example a given syndrome is associated with uncinate disconnection, many analyses could also show intersection with ILF and IFOF given they are larger and may give the impression that the syndrome is associated with disconnection of those tracts. Alternatively, if lesions were randomly distributed throughout the brain and you ran an analysis of that hypothetical syndrome you may reach the conclusion that intersection with larger tracts was associated with the syndrome given they are larger and more likely to intersect with lesions but the location might not in essence matter.
Thank you so much for your thoughts,
Isaiah
Sorry, Isaiah, your message was during my holiday, and I forgot to reply when I came back.
This is a good question. However bigger tracts are also more likely to be hit by lesions in patients that do not show your deficit of interest so I would still think that tract size is not a nuisance covariate.
cheers
mich
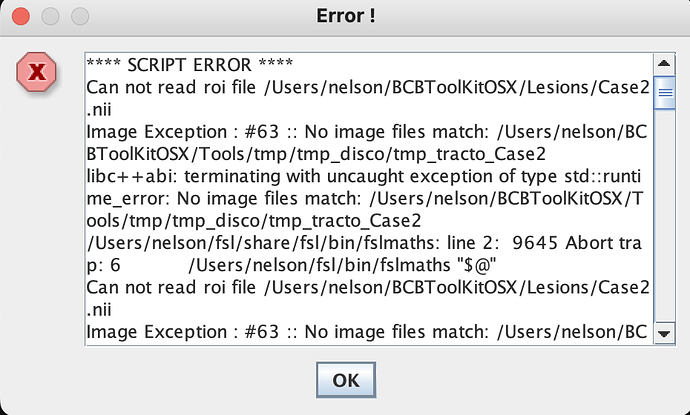
Hello,
I am using BCBtoolkit for disconnectome maps for the first time and I am running into an error. I’m upload a 2D binary lesion into the “lesion folder” and when I run the map I get the following message:
Can you help?
Thanks
Best,
Nelson
Hi Nelson,
It looks like there is an issue with your lesion file.
Is it in the MNI152? can you share this file with us.
More information on how to use BCBtoolkit can be found in the BCBtoolkit manual available at www.bcblab.com
cheers
mich
Hi,
I have two questions regarding the tractotron tool.
- The supplementary JHU atlas you provide in the software package has many abbreviations. Where can I find the full tract names of the JHU atlas that match these abbreviations?
- I found a white-matter tract from another atlas which I would like to investigate its disconnections with a dataset of lesion masks. The nifti file of this tract is a binary mask. The tractotron results were all nan values in probability.xsl and all 0 in proportion.xsl. Any way that tractotron can work with binary tract masks?
Thanks so much in advance!