Hello humans!
I am running BIDS validation on Healthy Brain Network (HBN) MRI data.
Site- subjects have data as:
<…>_FLAIR.json
<…>_FLAIR.nii.gz
<…>_MT-off_PD.json
<…>_MT-off_PD.nii.gz
<…>_MT-on_PD.json
<…>_MT-on_PD.nii.gz
<…>_T1w.json
<…>_T1w.nii.gz
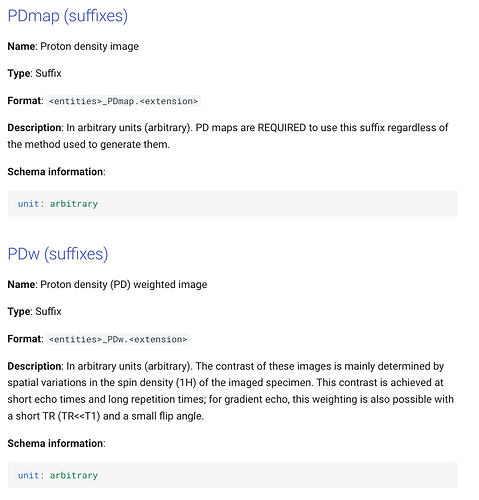
According to BIDS website:
I am confused which suffix should i use instead of _PD - PDw or PDmap?
Here’s _PD.json content example:
{
"InPlanePhaseEncodingDirectionDICOM": "ROW",
"ProcedureStepDescription": "CMI_HBN",
"DeviceSerialNumber": "26121",
"AcquisitionMatrixPE": 256,
"ImageOrientationPatientDICOM": [
0.0261769,
0.999657,
1.918e-08,
0.059286,
-0.00155244,
-0.99824
],
"ManufacturersModelName": "Avanto",
"ProtocolName": "MT_OFF_-SAG",
"RepetitionTime": 0.03,
"MagneticFieldStrength": 1.5,
"PhaseEncodingSteps": 191,
"MRAcquisitionType": "3D",
"ParallelReductionFactorInPlane": 2,
"SliceThickness": 1,
"DwellTime": 5.6e-06,
"TxRefAmp": 236.752,
"SAR": 0.0068763,
"ConversionSoftware": "dcm2niix",
"ScanningSequence": "GR",
"Manufacturer": "Siemens",
"PercentPhaseFOV": 100,
"ReconMatrixPE": 256,
"FlipAngle": 15,
"SeriesDescription": "MT_OFF_-SAG",
"PartialFourier": 0.75,
"ConversionSoftwareVersion": "v1.0.20171204 (OpenJPEG build) GCC4.8.4",
"ShimSetting": [
95,
-151,
197,
325,
391,
-24,
-168,
362
],
"PulseSequenceDetails": "%SiemensSeq%_gre",
"PatientPosition": "HFS",
"SequenceName": "_fl3d1",
"ReceiveCoilName": "32Ch_Head",
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"ND"
],
"EchoTime": 0.011,
"SequenceVariant": "SP",
"PhaseResolution": 1,
"BaseResolution": 256,
"PixelBandwidth": 349,
"ScanOptions": "PFP",
"SoftwareVersions": "syngo_MR_B17",
"StationName": "MRC26121",
"Modality": "MR"
}