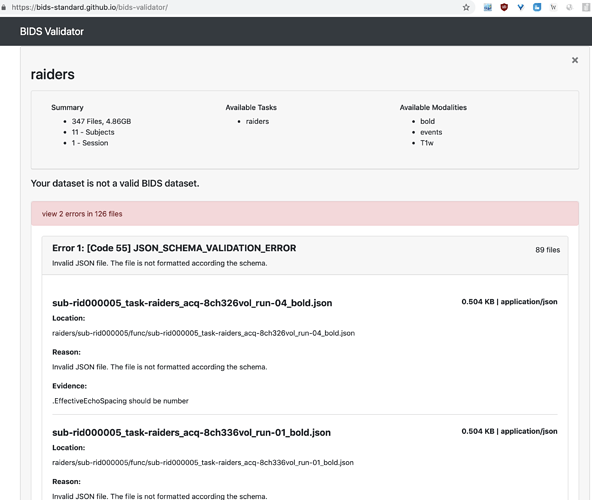
Hi, I get this error for all the JSON files in the dataset, however I’m fairly certain I do not need EffectiveEchoSpacing for my task, can I still go ahead and preprocess the data with these errors?
The dataset is http://datasets.datalad.org/?dir=/labs/haxby/raiders.
A few things:
-
EffectiveEchoSpacingmight be necessary to perform susceptibility distortion correction. - bids-validator tests are not independent. This error might prevent other tests to run and thus hide other issues with the dataset.
- For the sake of you down the road and your collaborators, it is always good to strive to improve the quality of dataset curation. This means fixing this issue. IF you are not the person who distributes the dataset it might be worth reaching to them and reporting this (perhaps @yarikoptic knows who to get in touch?)
Oops – sorry about that placeholder value (“TODO”) left behind. Removed now. git pull or datalad update --merge to fetch the fix. Here is the fix:
git show 33b78957c0beec524f077ecc6c62736c2f4af407
commit 33b78957c0beec524f077ecc6c62736c2f4af407
Author: Yaroslav Halchenko <debian@onerussian.com>
Date: Thu Feb 28 13:06:03 2019 -0500
Removed EffectiveEchoSpacing with bogus 'TODO' placeholder for the value
See https://neurostars.org/t/bids-validator-error-code-55-json-schema-validation-error-effectiveechospacing/3704
diff --git a/.datalad/metadata/objects/d2/ds-2b8160f327300514ee6915a75b5f19 b/.datalad/metadata/objects/d2/ds-2b8160f327300514ee6915a75b5f19
index 0da415b4..de50fa6e 100644
--- a/.datalad/metadata/objects/d2/ds-2b8160f327300514ee6915a75b5f19
+++ b/.datalad/metadata/objects/d2/ds-2b8160f327300514ee6915a75b5f19
@@ -276,10 +276,6 @@
0.00453,
0.035
],
-"EffectiveEchoSpacing":
-[
-"TODO"
-],
"FlipAngle":
[
8,
@@ -841,4 +837,4 @@ null,
}
}
}
-}
\ No newline at end of file
+}
diff --git a/task-raiders_bold.json b/task-raiders_bold.json
index 5226e4d8..fbec927a 100644
--- a/task-raiders_bold.json
+++ b/task-raiders_bold.json
@@ -1 +1 @@
-{"EchoTime": 0.035, "RepetitionTime": 2.5, "MagneticFieldStrength": 3, "dcmmeta_slice_dim": "TODO", "FlipAngle": 90, "ProcedureStepDescription": "Haxby_Nastase_raiders", "CogAtlasID": "http://www.cognitiveatlas.org/task/film_viewing", "PhaseEncodingDirection": "j-", "EffectiveEchoSpacing": "TODO", "dcmmeta_reorient_transform": [[0.0, -1.0, 0.0, 95.0], [1.0, 0.0, 0.0, 0.0], [0.0, 0.0, 1.0, 0.0], [0.0, 0.0, 0.0, 1.0]], "ManufacturersModelName": "Achieva", "dcmmeta_version": 0.6, "TaskName": "raiders", "dcmmeta_shape": [80, 80, 41], "ImageType": ["ORIGINAL", "PRIMARY", "FMRI", "NONE", "ND", "NORM", "MOSA"], "Manufacturer": "Philips"}
\ No newline at end of file
+{"EchoTime": 0.035, "RepetitionTime": 2.5, "MagneticFieldStrength": 3, "dcmmeta_slice_dim": "TODO", "FlipAngle": 90, "ProcedureStepDescription": "Haxby_Nastase_raiders", "CogAtlasID": "http://www.cognitiveatlas.org/task/film_viewing", "PhaseEncodingDirection": "j-", "dcmmeta_reorient_transform": [[0.0, -1.0, 0.0, 95.0], [1.0, 0.0, 0.0, 0.0], [0.0, 0.0, 1.0, 0.0], [0.0, 0.0, 0.0, 1.0]], "ManufacturersModelName": "Achieva", "dcmmeta_version": 0.6, "TaskName": "raiders", "dcmmeta_shape": [80, 80, 41], "ImageType": ["ORIGINAL", "PRIMARY", "FMRI", "NONE", "ND", "NORM", "MOSA"], "Manufacturer": "Philips"}
Re bids-validator - I am a bit confused since it complains about subject specific _bold.json whenever the bug was at the top level common task- file. I have checked with currently available from docker (via singularity) 0.21.1 and it correctly reports the problem in that task-raiders_bold.json. Unfortunately dunno what is the version of validator online, so filed and issue
As for fixing it properly (i.e. specifying the value) – not sure if we have verifiable source for that metadata any longer – that scanner (Philips) was decommissioned 3 years ago. You can find the original PAR/REC files (no DICOMs were exported - only PAR/REC and NIfTIs) if you checkout openfmri-1.0.0 tag, and PAR files might provide some additional metadata on top what we have in .json’s already, but I do not think we have anything to obtain EffectiveEchoSpacing any longer.
That’s weird the latest version available on Docker Hub is 1.1.4 see https://hub.docker.com/r/bids/validator/tags (the actual latest version is 1.1.9, but it’s not available on Docker, due to a deployment bug that was fixed - next version should end up there when it is released).
heh – I was using docker://bids/base_validator . Will check tomorrow and will whine if reproduce since, I guess due to all the inheritance, validator (in the webshot) somewhat incorrectly imho points to the cause of the trouble - that JSON file is a valid one. It is a json which validator dynamically composes via inheritance - it is the one invalid. Ideally I want to be pointed to the location of the error, but realizing that it might be difficult – at least the message could be a bit not misleading/more informative
Hello communtiy,
we are using docker, singularity and heudiconv to create a BIDS compatible dataset.
Unfortunately the BIDS validator gives the following error:
JSON_SCHEMA_VALIDATION_ERROR
sub-01_ses-005_task-rest_run-001_bold.json43.582 KB
Location:
Nifti/sub-01/ses-005/func/sub-01_ses-005_task-rest_run-001_bold.json
Reason:
Invalid JSON file. The file is not formatted according the schema.
Evidence:
.CogAtlasID should match format “uri”
task-rest_bold.json43.631 KB
Location:
Nifti/task-rest_bold.json
Reason:
Invalid JSON file. The file is not formatted according the schema.
Evidence:
.CogAtlasID should match format “uri”
I am not sure how to solve that issue?
I checked the json file with a json validator, the file itself is perfectly fine so I assume it is something about the Dicom header and the info that is saved in the json file? We have T1 images and resting state BOLD.
Thank you for your help.
Carina
Thank you for your message. The error is flagging the CogAtlasID field entry in those json files - can you please confirm the value is a uri
Thank you,
Franklin
Hi Franklin,
thank you for your reply.
There is no entry in the CogAtlasID field. I can delete that field and it is a valid BIDS folder, but I am not sure if this is an important field for later preprocessing (fMRIprep).
I think I don’t understand what the doi in that field would tell me.
Thank you so much.
Carina
It is not used in fMRIprep and I don’t think there is any type of processing that relies on it. The intent of the field was to provide a space to link to a description in an ontology of the task being performed (if it had one) to give it more context.
yep! As Ross mentioned this field is simply utilized for linking to further information about your dataset. If this field is not being used, it can be removed from your json file.
Thank you,
Franklin
Hi folks,
I’m getting the same error. Although none of my json sidecars have the “CogAtlasID” to begin with so I’m not sure why I get the following error for all 22 subjects in the dateset with functional scans
./sub-004/ses-01/func/sub-004_ses-01_task-motor_run-01_bold.json
Evidence: .CogAtlasID should match format "uri"
It gets resolved if I add a “CogAtlasID” field with a uri string. But since this only a recommended field and not a required one, I was wondering why there’s an error.
hi @Arsh
Thank you for surfacing you are running into this too. That’s odd it is raising this even though no CogAtlasID's are listed in your sidecar files.
@rwblair perhaps may have thoughts on why this message is appearing
Thank you,
Franklin
Hi everyone,
I’m running into this same error:
Error 1: [Code 55] JSON_SCHEMA_VALIDATION_ERROR
Invalid JSON file. The file is not formatted according the schema.1 file
task-flanker_bold.json 0.878 KB | application/json
Location:
Flanker/task-flanker_bold.json
Reason:
Invalid JSON file. The file is not formatted according the schema.
Evidence:
.CogAtlasID should match format “uri”
When I go into task-flanker_bold.json, this is what I can see: “CogAtlasID”: “tsk_4a57abb949a4f”,
Does anyone know how to rectify this?
Many thanks!
Phoenix
I believe it wants a URI like http://cognitiveatlas.org/task/id/tsk_4a57abb949a4f/ instead of just the id.
Thank you for your response, I took your suggestion and I’m still experiencing the same problem?
I ran into this same error regarding the CogAtlasID field when helping a trainee upload a dataset to OpenNeuro.
You might check the task-<taskname>_bold.json file that sits at the top level of your BIDS directory. That was the only json file that had the CogAtlasID field (default heudiconv value = “TODO”). Once we deleted that line from the task-<taskname>_bold.json file, the CogAtlasID error went away.
cheers.
suzanne